TAXONOMY—THE VOCABULARY OF MEDICAL MICROBIOLOGY
One has only to peruse the table of contents of this book to appreciate the diversity of medical pathogens that are associated with infectious diseases. It has been estimated that we currently have the capacity to identify fewer than 10% of the pathogens responsible for causing human disease. This is due to our inability to culture or target these organisms using molecular probes. The diversity of even these identifiable pathogens alone is so great that it is important to appreciate the subtleties associated with each infectious agent. The reason for understanding these differences is significant because each infectious agent has specifically adapted to a particular mode(s) of transmission, a mechanism(s) to grow in a human host (colonization), and a mechanism(s) to cause disease (pathology). As such, a vocabulary that consistently communicates the unique characteristics of infectious organisms to students, microbiologists, and health care workers is critical to avoid the chaos that would ensue without the organizational guidelines of bacterial taxonomy (Gk. taxon = arrangement; eg, the classification of organisms in an ordered system that indicates a natural relationship).
Identification, classification, and nomenclature are three separate but interrelated areas of bacterial taxonomy. Each area is critical to the ultimate goal of accurately studying the infectious diseases and precisely communicating these to others in the field.
Identification is practical use of a classification scheme to (1) isolate and distinguish specific organisms among the mix of complex microbial flora, (2) verify the authenticity or special properties of a culture in a clinical setting, and (3) isolate the causative agent of a disease. The latter may lead to the selection of specific pharmacologic treatments directed toward their eradication, a vaccine mitigating their pathology, or a public health measure (eg, handwashing) that prevents further transmission.
Identification schemes are not classification schemes, although there may be some superficial similarity. For example, the popular literature has reported Escherichia coli as being a cause of hemolytic uremic syndrome (HUS) in infants. There are hundreds of different strains that are classified as E coli but only a few that are associated with HUS. These strains can be “identified” from the many other E coli strains by antibody reactivity with their O-, H-, and K-antigens, as described in Chapter 2 (eg, E coli O157:H7). However, they are more broadly classified as a member of the family Enterobacteriaceae.
In a microbiologic context, classification is the categorization of organisms into taxonomic groups. Experimental and observational techniques are required for taxonomic classification. This is because biochemical, physiologic, genetic, and morphologic properties are historically necessary for establishing a taxonomic rank. This area of microbiology is necessarily dynamic as the tools continue to evolve (eg, new methods of microscopy, biochemical analysis, and computational nucleic acid biology).
Nomenclature refers to the naming of an organism by an established group of scientific and medical professionals. This is arguably the most important component of taxonomy because it allows medical professionals to communicate with each. Just as our societal vocabulary evolves, so does the vocabulary of medical microbiology. Any professional associated with infectious disease should be aware of the evolving taxonomy of infectious microorganisms.
Ultimately, the taxonomic ranks form the basis for the organization of bacteria. Linnaean taxonomy is the system most familiar to biologists. It uses the formal ranks of kingdom, phylum, class, order, family, genus, and species. The lower ranks are approved by a consensus of experts in the scientific community. Of these ranks, the family, genus, and species are the most useful (Table 3-1).
CRITERIA FOR IDENTIFICATION OF BACTERIA
Suitable criteria for bacterial classification include many of the properties that were described in the preceding chapter. One criterion is growth on different types of bacteriologic media. The general cultivation of most bacteria requires media rich in metabolic nutrients. These media generally include agar, a carbon source, and an acid hydrolysate or enzymatically degraded source of biologic material (eg, casein). Because of the undefined composition of the latter, these types of media are referred to as complex media.
Clinical samples from normally nonsterile sites (eg, the throat or the colon) contain multiple species of organisms, including potential pathogens and resident microbial flora. Media can be nonselective or selective; the latter are used to distinguish among the various bacteria in a clinical sample containing many different organisms.
Blood agar and chocolate agar are examples of complex, nonselective media, which support the growth of many different bacteria. These media are intended to cultivate as many species as possible, thus giving rise to numerous types of bacterial colonies.
Because of the diversity of microorganisms that typically reside at some sampling sites (eg, the skin, respiratory tract, intestines, vagina), selective media are used to eliminate (or reduce) the large numbers of irrelevant bacteria in these specimens. The basis for selective media is the incorporation of an inhibitory agent that specifically selects against the growth of irrelevant bacteria. Examples of such agents are:
Sodium azide—selects for gram-positive bacteria over gram-negative bacteria
Bile salts (sodium deoxycholate)—select for gram-negative enteric bacteria and inhibit gram-negative mucosal and most gram-positive bacteria
Colistin and nalidixic acid—inhibit the growth of many gram-negative bacteria
Examples of selective media are MacConkey agar (contains bile) that selects for the Enterobacteriaceae and CNA blood agar (contains colistin and nalidixic acid) that selects for staphylococci and streptococci.
Upon culture, some bacteria produce characteristic pigments, and others can be differentiated on the basis of their complement of extracellular enzymes; the activity of these enzymes often can be detected as zones of clearing surrounding colonies grown in the presence of insoluble substrates (eg, zones of hemolysis in agar medium containing red blood cells).
Many of the members of the Enterobacteriaceae can be differentiated on the basis of their ability to metabolize lactose. For example, pathogenic salmonellae and shigellae that do not ferment lactose on a MacConkey plate form white colonies, while lactose-fermenting members of the Enterobacteriaceae (eg, E coli) form red or pink colonies.
The number of differential media used in today’s clinical laboratories is far beyond the scope of this chapter.
Historically, the Gram stain, together with visualization by light microscopy, has been among the most informative methods for classifying the eubacteria. This staining technique broadly divides bacteria on the basis of fundamental differences in the structure of their cell walls (see Chapter 2). This typically represents the first step in identifying individual microbial specimens (eg, are they gram negative or gram positive) grown in culture or even directly from patient specimens (eg, urine specimens).
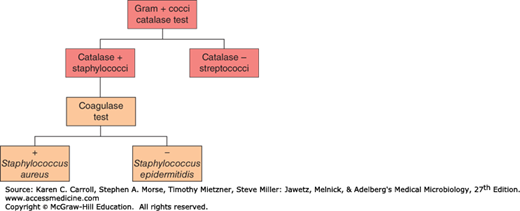
Tests such as the oxidase test, which uses an artificial electron acceptor, can be used to distinguish organisms on the basis of the presence or absence of a respiratory enzyme, cytochrome C, the lack of which differentiates the Enterobacteriaceae from other gram-negative rods. Similarly, catalase activity can be used, for example, to differentiate between the gram-positive cocci; the species staphylococci are catalase positive, whereas the species streptococci are catalase negative. If the organism is demonstrated to be catalase positive (Staphylococcus spp.), the species can be subdivided by a coagulase test into Staphylococcus aureus (coagulase positive) or Staphylococcus epidermitidis (coagulase negative) as demonstrated in Figure 3-1.
Ultimately, there are many examples of biochemical tests that can ascertain the presence of characteristic metabolic functions and be used to group bacteria into a specific taxon. A nonexhaustive list of common biochemical tests is given in Table 3-2.
|
The designation “sero” simply indicates the use of antibodies (polyclonal or monoclonal) that react with specific bacterial cell surface structures such as lipopolysaccharide (LPS), flagella, or capsular antigens. The terms “serotype,” “serogroups,” and “serovars” are, for all practical purposes, identical—they all use the specificity of these antibodies to subdivide strains of a particular bacterial species.
Under certain circumstances (eg, an epidemic), it is important to distinguish among strains of a given species or to identify a particular strain. This is called subtyping and is accomplished by examining bacterial isolates for characteristics that allow discrimination below the species level. Classically, subtyping has been accomplished by biotyping, serotyping, antimicrobial susceptibility testing, and bacteriophage typing. For example, more than 130 serogroups of Vibrio cholerae have been identified on the basis of antigenic differences in the O-polysaccharide of their LPS; however, only the O1 and O139 serogroups are associated with epidemic and pandemic cholera. Within these serogroups, only strains that produce a particular toxin-coregulated pili and cholera toxin are virulent and cause the disease cholera. By contrast, nontoxigenic V cholerae strains, which are not associated with epidemic cholera, have been isolated from environmental specimens, from food, and from patients with sporadic diarrhea.
Clonality with respect to isolates of microorganisms from a common source outbreak (point source spread) is an important concept in the epidemiology of infectious diseases. Etiologic agents associated with these outbreaks are generally clonal; in other words, they are the progeny of a single cell and thus, for all practical purposes, are genetically identical. Thus, subtyping plays an important role in discriminating among these particular microorganisms. Recent advances in biotechnology have dramatically improved our ability to subtype microorganisms. Hybridoma technology has resulted in the development of monoclonal antibodies against cell surface antigens, which have been used to create highly standardized antibody-based subtyping systems that describe bacterial serotypes. This is an important tool for defining the epidemiologic spread of a bacterial infection.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree