Overview
The nucleus is often represented as a relatively empty structure, containing only deoxyribonucleic acid (DNA) being replicated and transcribed along with a few accessory molecules to help in the process. To the contrary, the nucleus is actually a highly organized, membrane-bound structure that is literally filled with proteins, nucleotides, carbohydrates, and lipids with multiple functions. Various proteins are involved, along with the nuclear membrane, in the organization of chromosomes, which also helps to regulate the processes of DNA replication and transcription, and, subsequently, protein synthesis. Other proteins directly influence the expression of genes via direct interactions with specific nucleotide sequences. Posttranslational modifications affect both protein function and direct particular proteins to intracellular and/or extracellular destinations.
Structure of the Nucleus
The nucleus is surrounded by a double membrane called the nuclear membrane (also known as the nuclear envelope), with the outer layer being continuous with the endoplasmic reticulum (ER) in the cytoplasm and, like the ER, containing ribosomes and newly synthesized proteins. The inner and outer nuclear membranes are also continuous at the sites of nuclear pores. Approximately 2000 nuclear pores are contained within the nuclear membrane, and each pore can allow movement of about 1000 molecules in and out of the nucleus per second. These nuclear pores, composed of proteins called nucleoporins, are directly analogous to the membrane channels discussed in Chapter 8 and transport several types of molecules, including ribonucleic acid (RNA) and ribosomes, proteins, carbohydrates, and lipids. Smaller molecules and ions pass through the nuclear pore by simple diffusion, but larger proteins and RNA molecules are blocked by a spoke-like gate inside the channel and must be actively assisted by carrier proteins called “importins” or “exportins” by a process that requires two guanosine triphosphate (GTP) molecules. Each type of RNA molecule [messenger RNA (mRNA) and transfer RNA (tRNA)] that must be transported into the cytoplasm has an exportin and a specific amino acid (AA), nuclear export sequence, which directs them to a specialized nuclear pore for their selected transport out of the nucleus.
Leptomycins: Leptomycins A and B, originally developed as antifungal drugs, specifically alkylate an importin, which results in inhibition of the nuclear export of several RNAs and transcription regulators in cell cycle control. An additional leptomycin is HIV-1 “regulator, which allows HIV to take over host protein synthesis.” The leptomycins also stabilize p53, known to suppress tumor development/growth. Because of their cell cycle effects (see below), leptomycins are now being considered for cancer therapy. |
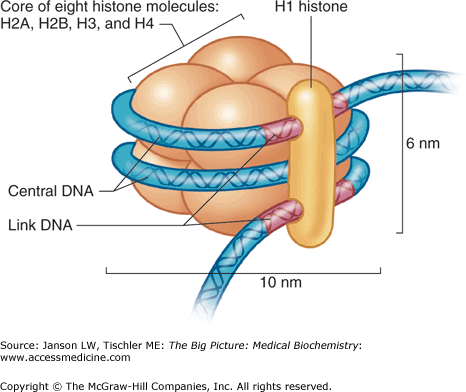
The most important structure inside the nucleus is chromatin, consisting, in humans, of the 46 chromosomes and their associated proteins. These proteins enable not only efficient packing of over 12 billion nucleotides in human DNA, but also selective unwinding of these chromosomes to expose genes for DNA replication, DNA to RNA transcription and mRNA processing. Histones are one major example of these associated proteins, and are separated into six classes: H1, H2A, H2B, H3, H4, and H5. Two each of histones H2A, H2B, H3, and H4 assemble into an eight-subunit nucleosome and wrap 146 or 147 DNA base pairs of the simple, double helix structure (Figure 9-1) around the complex.
Figure 9-1.
Histone Structure. The histone contains a core of histone molecules, including pairs of H2A, H2B, H3, and H4, wrapped by double-helix DNA and held together by histone H1. See text for further details. DNA, deoxyribonucleic acid. [Reproduced with permission from Mescher AL: Junqueira’s Basic Histology Text and Atlas, 12th edition, McGraw-Hill, 2010.]
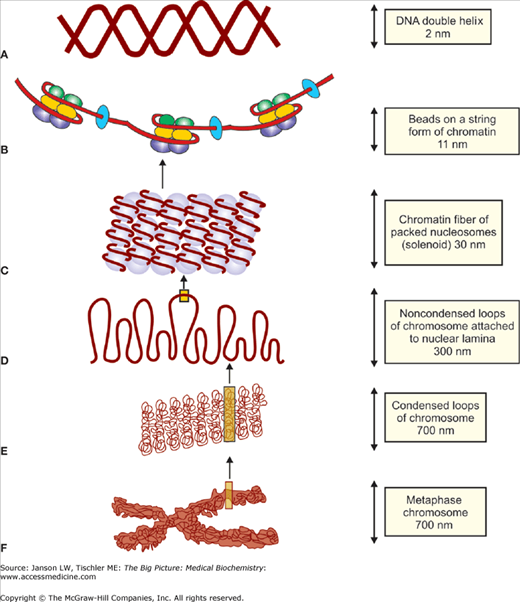
Several of these nucleosomes, linked together by about 50 DNA base pair sequences (Figure 9-2A), create an approximately 11-nm “beads on a string” chromatin structure (Figure 9-2B). Addition of the H1 histone (Figure 9-1) covers the entry and exit points of the DNA molecule and allows DNA to form coiled-coil structures, including the 30-nm chromatin fiber (Figure 9-2C). The 30-nm fiber can readily unwind into its 11-nm component to allow DNA replication and transcription. Analogous proteins called protamines are found in sperm and allow even denser packing of DNA in the sperm head than histones.
Figure 9-2.
A-F. DNA Structure from Chromosome to Double-Helix. Initial DNA structure beyond the simple, double helix (A) results from histones H2A, H2B, H3, and H4 interactions with DNA to create (B) 11-nm “beads on a string” structure. (C) The subsequent addition of histone H1 creates the 30-nm fiber. Further tertiary and quaternary structures of DNA are created by the addition of various scaffolding proteins, which condense DNA into varying compact chromosome structures. The level of structure varies depending on the cell cycle stage and, as a result, the requirement for DNA transcription or replication [e.g, (D) noncondensed, interphase structures (300 nm) and (E) condensed, metaphase structure (700 nm), ending with (F) the highly compacted structure of the metaphase chromosome]. DNA, deoxyribonucleic acid. [Adapted with permission from Naik P: Biochemistry, 3rd edition, Jaypee Brothers Medical Publishers (P) Ltd., 2009.]
Important protein attributes (Chapter 1) include positive/negative/uncharged AAs (primary structure), alpha helices (secondary structure), and grooves formed between separate histone proteins that provide and optimize essential interaction points between the histones and the DNA molecules. The properties of nucleotides also contribute to this binding because increased numbers of adenosine (A) and thymine (T) residues at the minor groove of DNA allow improved nucleosome binding. Not surprisingly, histone AA sequences and, therefore, higher order structures are highly conserved in biology. However, histones do much more than just wind DNA to a manageable size. Several histones are modified on their N-terminal tails and globular areas (see posttranslational modifications below). These modifications include the addition of methyl, acetyl, phosphate, ubiquitin, and adenosine diphosphate groups among others. The modifications change how the proteins interact with the DNA molecule, to expose (e.g., acetylation) and conceal (e.g., methylation) particular parts of the DNA strand as a part of activation and inhibition of gene transcription (gene regulation), as well as the repair of DNA errors from both DNA replication and ongoing processes that lead to nucleotide mutations.
Further compaction of the structure is provided by the nuclear matrix or “scaffold” proteins, which help in maintaining the structure and integrity of the DNA molecules. The exact structure and function of this structural element of the nucleus are still highly disputed. However, the existence is generally accepted of a supportive, highly dynamic network that produces both chromatin structure and, thereby, its functions by influencing the molecules with which it interacts. In corroboration of the concept of a nuclear matrix, “scaffold/matrix attachment elements” (S/MARs) have been proposed/identified in which specific DNA sequences would be able to connect to the scaffolding proteins. The existence and function of S/MARs is also now considered in biotechnology involved in gene therapy and modulation of DNA expression to modify disease states.
The nuclear matrix or lamina is a structure thought to be a three-dimensional network of intermediate filaments, a third type of structural protein (Chapter 12), as well as peripheral and integral membrane proteins attached to the inner lipid bilayer. Within the nucleus, additional chromatin structure is created from the scaffolding proteins by the formation of further supercoiled structures. These higher orders of structure include a 30-nm zigzag formation and a 100-nm fiber (not shown), as well as a 300-nm fiber (Figure 9-2D) with noncondensed and, therefore, accessible loops and a highly condensed 700-nm fiber (Figure 9-2E) that is part of the final metaphase chromatin structure (Figure 9-2F) found in cells. Other “scaffolding” proteins link the chromatin to the nuclear matrix (essential for chromosome movement during cell replication) and the cytoplasmic matrix to the nuclear membrane and internal nuclear structures. Of course, chromatin structure changes dramatically from interphase throughout DNA replication.
Laminopathies: More than 2000 variants of medical disorders of the nuclear membrane are known, many of which are caused by mutations that affect the proteins of the nuclear membrane or lamina. Each affects an otherwise important role of proteins involved in transport or structure of the membrane bilayer or its membrane proteins, which directly affect DNA replication and translation and the processes of protein translation and transport. These “laminopathies” have far-reaching effects, often resulting in childhood or adolescent disorders of skeletal and cardiac muscles, lipid, skin, nerves, and white blood cells, as well as cancers, diabetes, premature aging, and even early death. |
The nuclear matrix is not only actively involved in chromosome separation and cell division but also helps to maintain the required structure for each of these functions. The nuclear matrix creates a chromatin structure in the nucleus that is ordered and constrained in discrete territories that reflect specific functions, but which, at times of need such as mitosis, can alter dramatically while still maintaining control and organization. The result of the primary, secondary, tertiary, and quaternary folding of the DNA genome is the compaction of about 1.8 m of human DNA about 40,000-fold so it can fit inside a microscopic nucleus. Of course, this level of compaction changes throughout the cell cycle, being maximally compacted during mitosis and minimally compacted during interphase. Finally, changes in the DNA structure, including the marked condensation and expansion of chromatin, occur during the normal cell cycle.
Changing Nuclear Matrix in Cancer: Recent research has shown that early changes in many human cancers include a notable and measurable change in the proteins that make up the nuclear matrix. Although evaluation of these changes is still ongoing, the ability to detect these changes in the nuclear matrix may offer a reliable way to detect cancers at very early stages of the disease. Reversal of these changes may also offer clinicians, insights into prognosis and cures. |
Also found within the nucleus is the nucleolus, made of proteins and nucleic acids, where ribosomal RNA (rRNA) is produced and ribosomes are assembled for export into the cytoplasm. The protein components of ribosomes, often called “r-proteins,” are made in the cytoplasm and transported to the nucleolus via a connected network of nuclear membrane pores and nucleolar channels. rRNA is transcribed in the nucleus by RNA polymerases pol I, II, and III, and often methylated or shortened. The resulting rRNA sequences are targeted to the nucleolus where they join with the r-proteins to make the 40S and 60S subunits of mammalian ribosome, which is subsequently exported through a nuclear pore to the cytoplasm.
DNA Replication and Transcription
The conversion of information in the genetic code to functioning proteins and nucleic acids relies on the ability of the DNA to replicate itself and to transcribe its code to mRNA for use in protein synthesis. DNA replication results in very few mismatched nucleic acid pairs, with an approximate error rate of only one mismatched nucleic acid per 10 million nucleotides. The process of transcription also has mechanisms that reduce errors, but these are far less efficient than that seen in DNA replication. Both processes involve several proteins essential to the process as functional or regulatory elements.
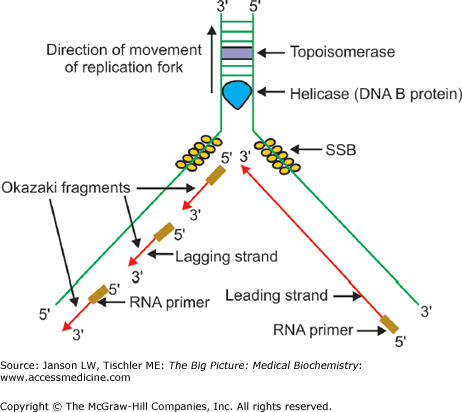
The process of uncoiling double-stranded DNA, faithfully copying each DNA strand and then separating the two, new, double-stranded copies, is called replication. The process starts at an origin of replication (ori), a particular sequence of nucleic acids at which a pre-replication complex (pre-RC) can bind and the replication can start. Approximately 100,000 origins of replication can be found in each human cell, allowing the copying of DNA to proceed in a parallel fashion from all of these points, thereby speeding the process of DNA replication. The pre-RC is composed of the following four proteins: (a) a six-subunit origin recognition complex binds first to the origin of replication; (b) two cell cycle regulatory proteins, Cdc6 and Cdt1, ensure that the cell is prepared for DNA replication; and (c) the minichromosome maintenance complex, which is believed to contain proteins essential for the establishment of a replication fork. The replication fork is the point where two DNA strands, one termed the leading strand and the other the lagging strand, are separated and DNA copying occurs. The coiled-coil, double-helical DNA structure examined in Chapter 4 is initially unwound by the enzyme DNA helicase (possibly part of the minichromosome maintenance complex) by breaking the hydrogen bonds between complementary nucleic acids. Single-stranded binding proteins attach to the new DNA strands to keep them separated. An enzyme termed primase then produces a short strand of RNA (sometimes with DNA) to serve as a primer for the remainder of the process. The enzyme DNA polymerase replicates each DNA strand in the 5′ to 3′ direction by adding the correct, matching nucleotide triphosphate to the 3′-hydroxyl end of the primer strand. As each new nucleic acid is added, a new phosphodiester bond is formed, utilizing the energy contained in the remaining diphosphate group. This process is continuous on the leading strand but, as DNA polymerase can only add in the 5′ to 3′ direction, short chains of newly added nucleic acids, called Okazaki fragments, are generated on the lagging strand. The enzyme DNA ligase joins the Okazaki fragments together as lagging strand replication proceeds. The process of replication along the coiled-coil structure of DNA soon leads to an unfavorable DNA conformation that is wound about itself. To relieve this problem, a DNA topoisomerase efficiently cuts the phosphate backbone, “untangles” the DNA strands, and then repairs the cut, leaving the DNA otherwise unaltered. This entire replication process is depicted in Figure 9-3.
Figure 9-3.
Overview of DNA Replication. Summary of DNA replication, illustrating the replication fork, leading and lagging strand synthesis, and the various proteins involved in replication and unwinding. A detailed description is offered in the text. DNA, deoxyribonucleic acid; RNA, ribonucleic acid. [Reproduced with permission from Naik P: Biochemistry, 3rd edition, Jaypee Brothers Medical Publishers (P) Ltd., 2009.]
Transcription is the process whereby genetic information from a DNA sequence is utilized to create an equivalent RNA—mRNA if the gene codes for a protein, tRNA for a tRNA, rRNA for assembly of a ribosome, or even catalytic RNA, termed ribozyme. This DNA sequence contains not only the gene for the RNA (coding sequence) but also regulatory sequences that dictate when and how the particular RNA will be produced. Again, the DNA is transcribed or “read” from 3′ to 5′ and occurs only on one of the DNA strands, known as the template strand.
Transcription starts with binding of the enzyme RNA polymerase to a promoter sequence on the DNA, usually located from 10 to 35 bases before the start of the actual gene (Figure 9-4
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree