1 The major tool in the study of histology is the microscope, and students will find a brief description of how microscopes work in Appendix 1 at the back of this book. Various staining procedures are used to prepare tissues to make them visible through the microscope and these are described in Appendix 2. The student is strongly urged to read these appendices first; it really will make all that follows more comprehensible. Appendix 3 is a glossary of common histological terms and, like the other appendices, can be dipped into at any time. FIG. 1.1 The cell (illustration opposite) FIG. 1.2 Membrane structure (illustrations opposite) The nucleus is the largest organelle in the cell and is usually the most obvious feature of the cell seen by light microscopy. The nucleus contains the genetic material of the cell, deoxyribonucleic acid (DNA), arranged in the form of chromosomes. Each chromosome contains a number of genes joined end to end, with each gene encoding the structure of a single protein according to the sequence of nucleotides along the length of the gene (see Fig. 1.6). Thus the genetic blueprint for all proteins, whether structural or enzymes, is contained within the nucleus of every cell in the body except for red blood cells, which have no nucleus (see Ch. 3). The substance of the nucleus is known as the nucleoplasm and it is surrounded by the nuclear membrane. The first step in cell division is replication of the DNA so that a copy of the genome goes to each of the daughter cells. Cell division is the subject of Ch. 2. FIG. 1.3 Nucleus (illustrations opposite) FIG. 1.4 Nuclear envelope FIG. 1.5 The nucleolus FIG. 1.6 Protein synthesis and degradation FIG. 1.7 Rough endoplasmic reticulum FIG. 1.8 Smooth endoplasmic reticulum
Cell structure and function
Introduction to the Cell
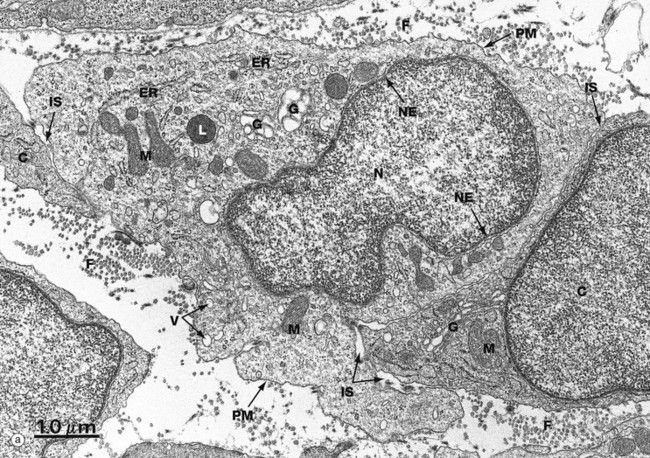
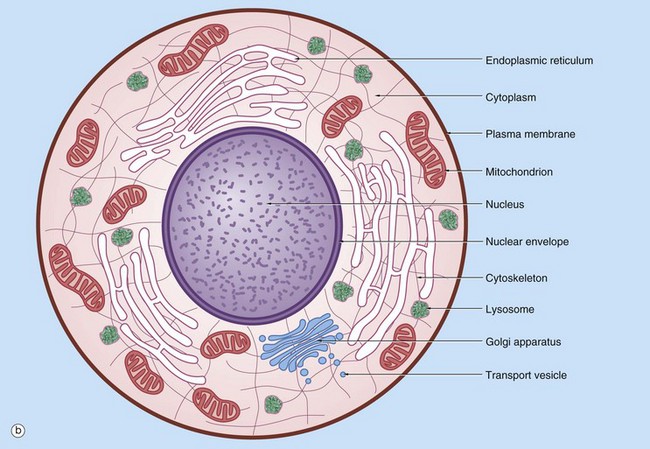
(a) EM ×16 500 (b) Schematic diagram
The basic structural features common to all eukaryotic cells are illustrated in this electron micrograph (a) of a fibroblast and diagram (b). All cells are bounded by an external lipid membrane, called the plasma membrane or plasmalemma PM, which serves as a dynamic interface with the external environment. Most cells interact with two types of external environment: adjacent cells C and extracellular matrix as represented by collagen fibrils F. The space between cells is designated the intercellular space IS. The functions of the plasma membrane include transfer of nutrients and metabolites, attachment of the cell to adjacent cells and extracellular matrix, and communication with the external environment.
The nucleus N is the largest organelle and its substance is bounded by a membrane system called the nuclear envelope or membrane NE. The nucleus contains the genetic material of the cell in the form of deoxyribonucleic acid (DNA). The cytoplasm contains a variety of other organelles, many of which are also bounded by membranes. An extensive system of flattened membrane-bound tubules, saccules and flattened cisterns, collectively known as the endoplasmic reticulum ER, is widely distributed throughout the cytoplasm. A second discrete system of membrane-bound saccules, the Golgi apparatus G, is typically located close to the nucleus (best seen in the adjacent cell). Scattered free in the cytoplasm are a number of relatively large, elongated organelles called mitochondria M, which have a smooth outer membrane and a convoluted inner membrane system. In addition to these major organelles, the cell contains a variety of other membrane-bound structures, including intracellular transport vesicles V and lysosomes L. The cytoplasmic organelles are suspended in a gel-like medium called the cytosol, in which many metabolic reactions take place. Within the cytosol, there is a network of minute tubules and filaments, collectively known as the cytoskeleton, which provides structural support for the cell and its organelles, as well as providing a mechanism for transfer of materials within the cell and movement of the cell itself.
Thus the cell is divided into a number of membrane-bound compartments, each of which has its own particular biochemical environment. Membranes therefore serve to separate incompatible biochemical and physiological processes. In addition, enzyme systems are found anchored in membranes so that membranes are themselves the site of many specific biochemical reactions. Membrane-enclosed compartments occupy approximately half the volume of the cell.
Membrane Structure
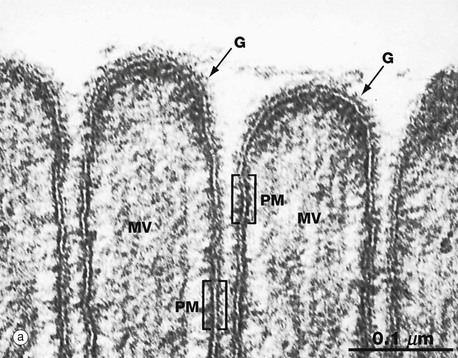
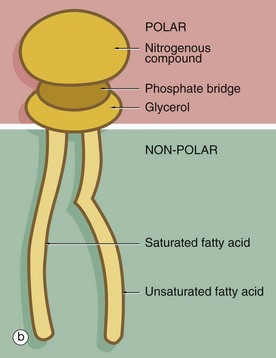
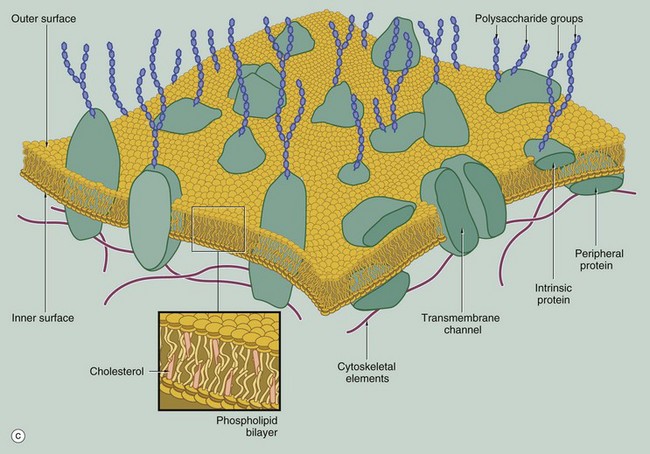
(a) EM ×210 000 (b) Phospholipid structure (c) Membrane structure
The phospholipid molecules that make up the lipid bilayers are amphiphilic, i.e. they consist of a polar, hydrophilic (water-loving) head and a non-polar, hydrophobic (water-hating) tail. Most often, the polar heads consist of glycerol conjugated to a nitrogenous compound such as choline, ethanolamine or serine via a phosphate bridge as shown in (b). The phosphate group is negatively charged, whereas the nitrogenous group is positively charged. The non-polar tail of the phospholipid molecule consists of two long-chain fatty acids, each covalently linked to the glycerol component of the polar head. In most mammalian cell membranes, one of the fatty acids is a straight-chain saturated fatty acid, while the other is an unsaturated fatty acid which is ‘kinked’ at the position of the unsaturated bond. Sphingomyelin is another important and plentiful phospholipid in cell membranes.
Phospholipids in aqueous solution will spontaneously form a bilayer with the hydrophilic (polar) heads directed outwards and the hydrophobic (non-polar) tails forced together inwards. The weak intermolecular (non-covalent) forces that hold the bilayer together allow individual phospholipid molecules to move freely within each layer, but exchange of lipids between the two layers is uncommon. The two lipid layers of the plasma membrane have different lipid composition and the lipid composition of the cell membrane is different in different cell types. The lipid structure of membranes is not homogeneous; certain lipids, glycolipids and proteins may be transiently enriched to form a membrane or lipid ‘raft’ which is involved in various membrane functions, including the formation of caveola (see Fig. 1.11).
The fluidity and flexibility of the membrane is increased by the presence of unsaturated fatty acids, which prevent close packing of the hydrophobic tails. Cholesterol molecules are also present in the bilayer in an almost 1 : 1 ratio with phospholipids. Cholesterol molecules themselves are amphiphilic and have a kinked conformation, thus preventing overly dense packing of the phospholipid fatty acid tails while at the same time filling the gaps between the ‘kinks’ of the unsaturated fatty acid tails. Cholesterol molecules thus stabilise and regulate the fluidity of the phospholipid bilayer.
As shown in diagram (c), protein molecules are embedded within the lipid bilayer (intrinsic or integral proteins). Some of these proteins span the entire thickness of the membrane (transmembrane proteins) to be exposed to each surface, while others are embedded within the inner or outer lipid leaflet. Membrane proteins are held within the membrane by a hydrophobic zone which allows the protein to move in the plane of the membrane. The parts of these proteins protruding beyond the lipid bilayer are hydrophilic. Some membrane proteins are anchored to cytoplasmic structures by the cytoskeleton. Peripheral membrane proteins are attached to the inner or outer membrane leaflet by weak non-covalent bonds to other proteins or lipids. Membrane proteins are important in cell-cell adhesion, cell-matrix adhesion, intercellular signalling and for the formation of transmembrane channels for transport of materials into and out of the cell. In many cases, the transmembrane proteins assemble into complexes of two or more protein molecules to form a transmembrane channel or signalling complex; one example is the aquaporins which transport water molecules across the cell membrane.
On the external surface of the plasma membranes of animal cells, most of the membrane proteins and some of the membrane lipids are conjugated with short chains of polysaccharide (carbohydrate); these glycoproteins (surface mucins) and glycolipids project from the surface of the bilayer forming an outer coating, the glycocalyx, which varies in thickness in different cell types. The glycocalyx is involved in cell recognition phenomena, in the formation of intercellular adhesions and in the adsorption of molecules to the cell surface; the glycocalyx also provides mechanical and chemical protection for the plasma membrane.
The electron micrograph in (a) provides a high-magnification view of the plasma membrane PM of the minute surface projections (microvilli) MV of a lining cell from the small intestine. The characteristic trilaminar appearance is made up of two outer electron-dense layers separated by an electron-lucent layer. The outer dense layers correspond to the hydrophilic heads of phospholipid molecules, while the electron-lucent layer represents the intermediate hydrophobic layer, mainly consisting of fatty acids and cholesterol. On the external surface of the plasma membrane, the glycocalyx G is seen as a fuzzy edge to the cell membrane. This is an unusually prominent feature of small intestinal lining cells where it incorporates a variety of digestive enzymes.
Plasma membranes mediate the flow of both materials and information into and out of the cell, a function of vital importance to the cell. This topic is dealt with in detail in the section ‘Import, export and intracellular transport’ later in this chapter.
The Nucleus
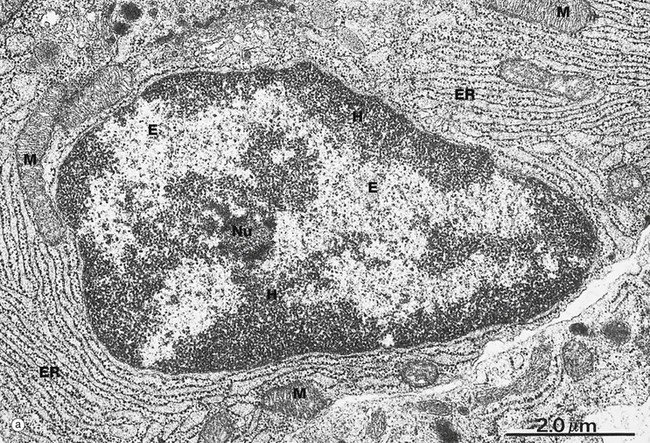
(a) EM ×15 000 (b) H&E (HP)
Micrograph (a) illustrates the nucleus of a plasma cell, a type of cell that secretes large amounts of a protein called antibody. Typical of protein-secreting cells, the cytoplasm contains plentiful ribosome-studded (rough) endoplasmic reticulum ER as well as mitochondria M, which produce the energy required for such a metabolically active cell.
The nucleus contains DNA (making up less than 20% of its mass), protein called nucleoprotein and some ribonucleic acid (RNA). Nucleoprotein is of two major types: low molecular weight, positively charged histone proteins and non-histone proteins. Histone proteins form a protein core around which the chromosome is coiled to form nucleosomes and control the uncoiling and expression of the genes encoded by the DNA strand. Non-histone proteins include all the enzymes for the synthesis of DNA and RNA and other regulatory proteins. All nucleoproteins are synthesised in the cytoplasm and imported into the nucleus. Nuclear RNA includes newly synthesised messenger, transfer and ribosomal RNA (mRNA, tRNA and rRNA, respectively) that has not yet passed into the cytoplasm. Control of DNA transcription is mediated by a variety of small RNA molecules including micro RNA (miRNA), small nuclear RNA (snRNA) and small interfering RNA (siRNA).
Except during cell division, the chromosomes, each a discrete length of DNA with bound histone proteins, exist as coiled and supercoiled strands that cannot be visualised individually. Nuclei are heterogeneous structures with electron-dense (dark, see App. 1) and electron-lucent (light) areas. The dense areas, called heterochromatin H, consist of tightly coiled inactive chromatin found in irregular clumps, often around the periphery of the nucleus. In females, the inactivated X-chromosome may form a small discrete mass, the Barr body. Barr bodies are occasionally seen at the edge of the nucleus in female cells when cut in a favourable plane of section. The electron-lucent nuclear material, called euchromatin E, represents that part of the DNA that is active in RNA synthesis. The nucleolus Nu is also evident (see Fig. 1.5). The name chromatin is derived from the strong colour of nuclei when stained for light microscopy. The chromatin is a highly organised but dynamic structure, with individual chromosomes tending to clump in particular areas of the nucleus, known as chromosome territories. Segments of the chromosome are coiled and uncoiled as different genes are brought into contact with the enzymes that make the RNA copy of the DNA, i.e. transcription. Histone proteins also exist as variant forms or can be chemically modified in ways that promote or suppress expression of a particular gene. Permanent switching on or off of a particular set of genes leads to differentiation of the cell.
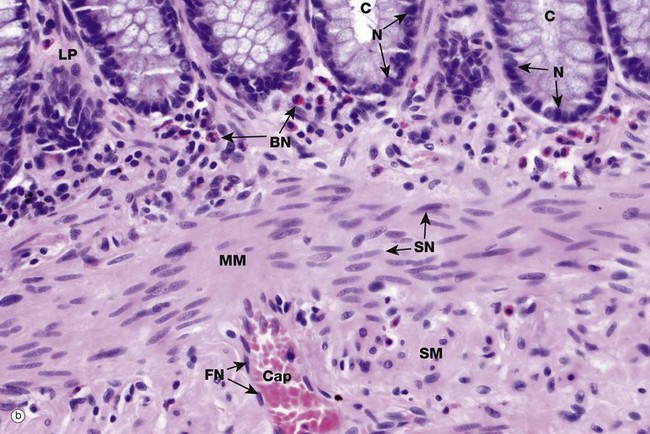
The shape, appearance and position of cell nuclei can be very helpful in identifying particular cell types. Micrograph (b) shows part of the wall of the colon (see also Fig. 14.29), which has been stained with haematoxylin and eosin (H&E, see Appendix 2), the ‘standard’ histological staining method. Haematoxylin is blue in colour and eosin is pink. Haematoxylin, a basic dye which binds to negatively charged DNA and RNA, stains nuclei dark blue. Eosin, an acidic dye, has affinity for positively charged structures such as mitochondria and many other cytoplasmic constituents and so the cytoplasm is stained pink. This micrograph shows the characteristic appearances of the nuclei of various cell types. At the top of the image, the bases of the colonic crypts C can be seen. The epithelial cells forming the crypts have round to ovoid nuclei N, typical of epithelial cells. Note also that the nuclei are positioned at the bases of the cells while the superficial cytoplasm is filled with mucin; the position of the nucleus within a cell, the polarity, may also be highly characteristic. Deep to the crypts, running across the centre of the image, is the muscularis mucosae MM, which is composed of smooth muscle cells. Smooth muscle cell nuclei SN are elongated with rounded ends, often called spindle shaped. This is of course only apparent if they are cut in the right plane of section; if cut perpendicular to the long axis of the cell, they appear rounded (see also Fig. 6.15). The nuclei are typically placed in the centre of the cell, although this is not always easy to see as the cell borders are indistinct. Eosinophils scattered within the lamina propria LP have a unique bilobed nuclear form BN which, together with the prominent coral red granules in the cytoplasm, makes identification easy. Note again that if the plane of section is unfavourable, the bilobed structure of the nucleus is not apparent. A very small blood vessel, a capillary Cap, is seen in the submucosa SM and the flattened nuclei FN of the endothelial cells can be identified; the cytoplasm of these cells is so thin that it cannot be seen at this magnification.
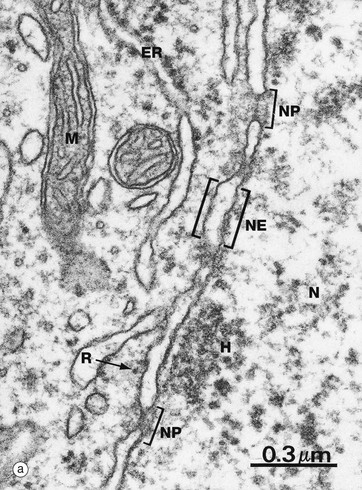
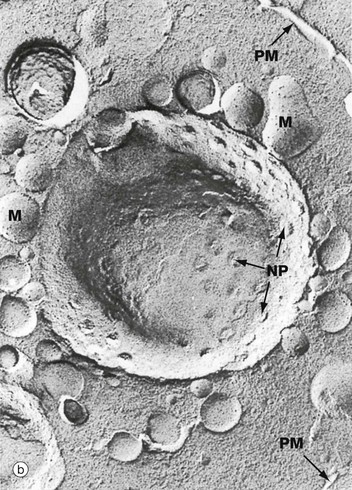
(a) EM ×59 000 (b) Freeze-etched preparation, SEM ×34 000
The nuclear envelope NE, which encloses the nucleus N, consists of two lipid bilayers with the intermembranous or perinuclear space between. The inner and outer nuclear membranes have the typical phospholipid bilayer structure but contain different integral proteins. The outer lipid bilayer is continuous with the endoplasmic reticulum ER and has ribosomes R on its cytoplasmic face. The intermembranous space is continuous with the lumen of the endoplasmic reticulum. On the inner aspect of the inner nuclear membrane, there is an electron-dense layer of intermediate filaments, the nuclear lamina, consisting of intermediate filaments called lamins that link inner membrane proteins and heterochromatin H.
The nuclear envelope contains numerous nuclear pores NP, at the margins of which the inner and outer membranes become continuous. Each pore contains a nuclear pore complex, an elaborate cylindrical structure consisting of approximately 30 proteins known as nucleoporins, forming a central pore approximately 125 nm in diameter. Nuclear pores permit and regulate the exchange of metabolites, macromolecules and ribosomal subunits between nucleus and cytoplasm. Ions and small molecules diffuse freely through the nuclear pore. Larger molecules, such as mRNA moving from nucleus to cytoplasm and histones from cytoplasm to nucleus, dock to the nuclear pore complex by means of a targeting sequence and are moved through the pore by an energy-dependent process. The nuclear pore complex may also hold together the two lipid bilayers of the nuclear envelope. Note that mitochondria M are also identifiable in the cytoplasm.
Micrograph (b) shows an example of a technique called freeze-etching. Briefly, this method involves the rapid freezing of cells which are then fractured. Internal surfaces of the cell are exposed at random, the fracture lines tending to follow natural planes of weakness. Surface detail is obtained by ‘etching’ or sublimating excess water molecules from the specimen at low temperature. A thin carbon impression is then made of the surface and this mirror image is viewed by scanning electron microscopy. Freeze-etching provides a valuable tool for studying internal cell surfaces at high resolution. In this preparation, the plane of cleavage has included part of the nuclear envelope in which nuclear pores NP are clearly demonstrated. Note also the outline of the plasma membrane PM and mitochondria M.
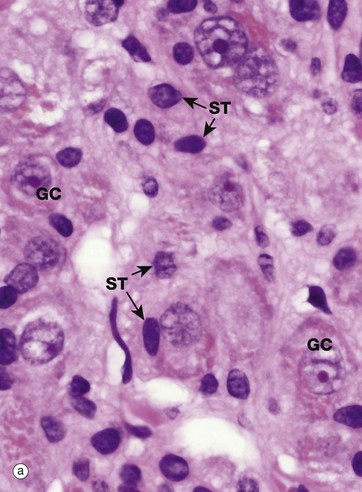
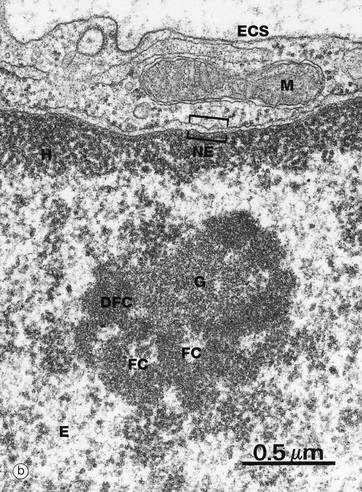
(a) H&E (HP) (b) EM ×37 000
Most nuclei contain a dense structure called the nucleolus, which is the site of ribosomal RNA (rRNA) synthesis and ribosome assembly. Transfer RNA (tRNA) is also processed in the nucleolus. More recently discovered roles include control of the cell cycle and stress responses. The nucleolus may be very prominent in some cell types and quite inconspicuous in others, as shown in micrograph (a) which is a high-power photomicrograph of an autonomic ganglion (see also Figs 7.21 and 7.22). In this micrograph, the nuclei of the ganglion cells GC contain large purple nucleoli, while the smaller sustentacular cell nuclei ST have small nucleoli that are only just visible at this magnification.
Furthermore, the nucleolus may change appearance depending on the state of the cell, so that an inactive fibroblast usually has a very small nucleolus while an activated fibroblast, for instance in a healing wound, has a prominent nucleolus. Remember that this is a thin slice of the tissue and that the plane of section does not go through the nucleolus of every cell, so that some nuclei appear to lack nucleoli.
The nucleolus is not membrane bound but consists of an aggregate of ribosomal genes, newly synthesised rRNA, ribosomal proteins and ribonucleoproteins. The ribosomal genes are found on five chromosomes and are called the nucleolar organiser regions (NORs). The rRNA is transcribed from the DNA template and then modified in the nucleolus and combined with ribosomal proteins. The subunits then pass back to the cytoplasm through the nuclear pore complex (NPC) to aggregate into complete ribosomes when bound to an mRNA molecule. Micrograph (b) is a high-power electron micrograph of a typical nucleolus. Nucleoli can be variable in appearance, but most contain dense fibrillar components DFC and paler fibrillar centres FC surrounded by the granular component G. The fibrillar components are the sites of ribosomal RNA synthesis, while ribosome assembly takes place in the granular components. Note also euchromatin E and heterochromatin H within the nucleus, which is bounded by the nuclear envelope NE. A thin rim of cytoplasm containing a mitochondrion M separates the nucleus from the extracellular space ECS.
Protein Synthesis
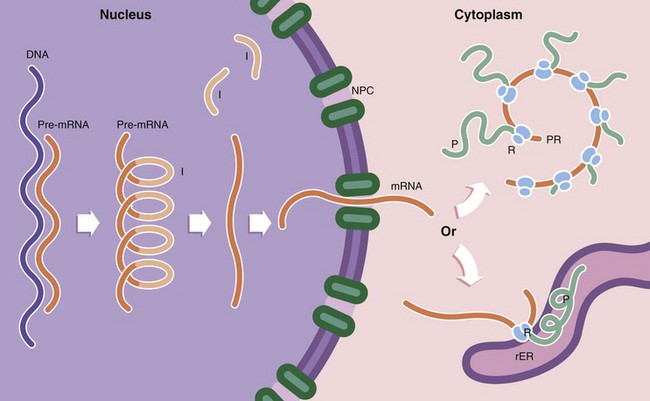
Protein synthesis occurs in several steps. First, the DNA template (the gene) of a particular protein is copied to form a complementary pre–messenger RNA (pre-mRNA) copy, a process known as transcription. Post-transcriptional processing of the mRNA results in excision of introns I (non-coding regions of the mRNA strand). This step is controlled by small nuclear RNAs (snRNA) which in combination with various proteins form the spliceosome. The messenger RNA (mRNA) then passes through the nuclear pore complex NPC into the cytoplasm. Here the mRNA binds to ribosomes R, organelles that synthesise proteins using the mRNA strand as a template to determine the specific amino acid sequence of the protein; this is known as translation. Ribosomes, which are synthesised in the nucleolus, comprise two subunits of unequal size. Each subunit consists of a strand of RNA, ribosomal RNA (rRNA) molecules, with associated ribosomal proteins forming a globular structure. Ribosomes align mRNA strands so that transfer RNA (tRNA) molecules may be brought into position and their amino acids added sequentially to the growing polypeptide chain P. Some of the RNA molecules in ribosomes catalyse peptide bond formation between amino acids and are sometimes called ribozymes to indicate this enzymatic function. Most enzymes are proteins. Thus the DNA code is converted first into RNA and then into a specific protein. Ribosomes are often found attached to mRNA molecules in small circular aggregations called polyribosomes or polysomes PR, formed by a single strand of mRNA with ribosomes attached along its length. Each ribosome in a polyribosome is making a separate molecule of the protein.
Ribosomes and polyribosomes may also be attached to the surface of endoplasmic reticulum. The ER consists of an interconnecting network of membranous tubules, vesicles and flattened sacs (cisternae) which ramifies throughout the cytoplasm. Much of its surface is studded with ribosomes, giving a ‘rough’ appearance, leading to the name rough endoplasmic reticulum rER. Proteins destined for export, as well as lysosomal proteins, are synthesised by ribosomes attached to the surface of the rER and pass through the membrane into its lumen. Integral membrane proteins are also synthesised on rER and inserted into the membrane at this point, the extracellular part of the protein protruding into the lumen of the rER and the intramembranous part held firmly in place by hydrophobic attraction. It is within the rER that many proteins are folded to form their tertiary structure, intrachain disulphide bonds are formed and the first steps of glycosylation take place. In contrast, proteins destined for the cytoplasm, nucleus and mitochondria are synthesised on free ribosomes and folding and other post-translational modifications take place there.
Proteins that are damaged or no longer required by the cell are degraded to short peptides. The first step in this process is binding of the protein ubiquitin to the damaged protein. This acts as a signal that allows the protein to be taken up by a proteasome. Proteasomes are non–membrane bound arrays of proteolytic enzymes that are plentiful in all cells. Other proteins are degraded by proteolytic enzymes within lysosomes.
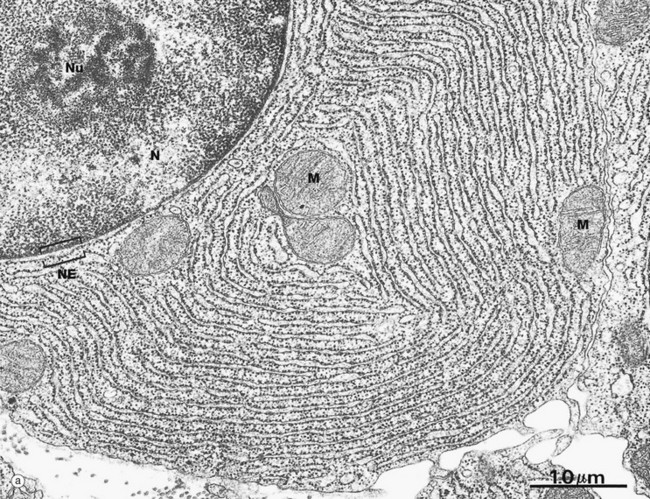
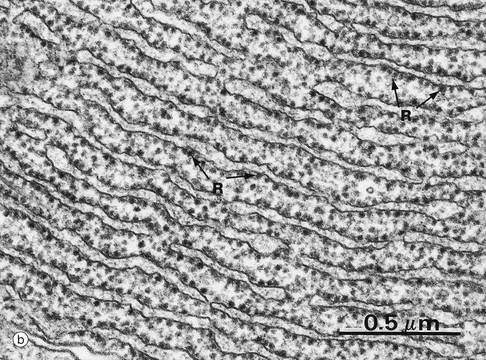
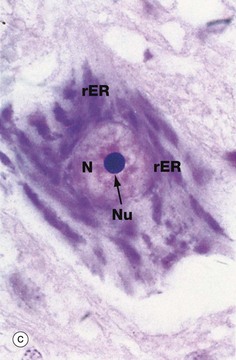
(a) EM ×23 000 (b) EM ×50 000 (c) Cresyl violet (HP)
These micrographs illustrate rough endoplasmic reticulum rER in a cell specialised for the synthesis and secretion of protein; in such cells, rER tends to be profuse and to form closely packed parallel laminae of flattened cisternae. In micrograph (a), the dimensions of the rER can be compared with that of mitochondria M and the nucleus N. The nucleus typically contains a prominent nucleolus Nu. Note the close association between the rER and the outer lipid bilayer of the nuclear envelope NE with which it is in continuity. The chromatin in the nucleus is mainly dispersed (euchromatin), consistent with prolific protein synthesis.
Micrograph (b) shows part of the rER at high magnification. Numerous ribosomes R stud the surface of the membrane system and there are plentiful ribosomes lying free in the intervening cytosol. Micrograph (c) shows a nerve cell at high magnification stained by the basophilic dye cresyl violet. The basophilic clumps in the cytoplasm represent areas of plentiful rER. The nuclear envelope can be distinguished due to the basophilia of the numerous ribosomes that stud its outer surface. The nucleus N contains a prominent nucleolus Nu and dispersed chromatin.
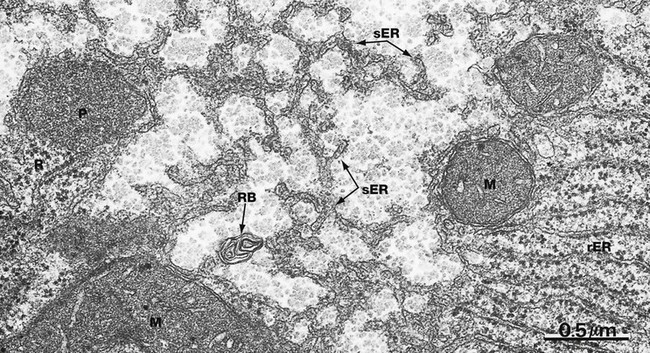
EM ×40 000
Smooth endoplasmic reticulum sER is continuous with and similar to rER except that it lacks ribosomes. The principal functions of smooth endoplasmic reticulum are lipid biosynthesis and membrane synthesis and repair. Fatty acids and triglycerides are mostly synthesised within the cytosol, whereas cholesterol and phospholipids are synthesised in sER. In liver cells, smooth endoplasmic reticulum is rich in cytochrome P450 and plays a major role in the metabolism of glycogen and detoxification of various noxious metabolic by-products, drugs and alcohol. In most cells, sER is involved in the storage and release of Ca2+ ions, an important mechanism of cell signalling. In muscle cells, where it is called sarcoplasmic reticulum, release and reuptake of Ca2+ ions activates the contractile mechanism (see Ch. 6).
Most cells contain only scattered elements of sER interspersed with the other organelles. Cell types with prominent sER include liver cells and those cells specialised for lipid biosynthesis, such as the steroid hormone-secreting cells of the adrenal glands and the gonads. In this micrograph from the liver, most of the membranous elements are sER, but it is continuous with rough endoplasmic reticulum rER in the lower right of the field. This field also includes several mitochondria M, a peroxisome P (see Fig. 1.24), free ribosomes and polyribosomes R and a whorl of membrane in a residual body RB (see Fig. 1.11).![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Basicmedical Key
Fastest Basicmedical Insight Engine
