Although any two homologous chromosomes generally look identical under the microscope, they differ substantially at the DNA sequence level. As discussed in Chapter 4, these differences at the same position (locus) on a pair of homologous chromosomes are alleles. Alleles that are common (generally considered to be those carried by approximately 2% or more of the population) constitute a polymorphism, and linkage analysis in families (as we will explore later in the chapter) requires following the inheritance of specific alleles as they are passed down in a family. Allelic variants on homologous chromosomes allow geneticists to trace each segment of a chromosome inherited by a particular child to determine if and where recombination events have occurred along the homologous chromosomes. Several tens of millions of genetic markers are available to serve as genetic markers for this purpose. It is a truism now in human genetics to say that it is essentially always possible to determine with confidence, through a series of analyses outlined in this chapter, whether a given allele or segment of the genome in a patient has been inherited from his or her father or mother. This advance—a singular product of the Human Genome Project—is an essential feature of genetic analysis to determine the precise genetic basis of disease.
Alleles at Loci on Different Chromosomes Assort Independently
Assume there are two polymorphic loci, 1 and 2, on different chromosomes, with alleles A and a at locus 1 and alleles B and b at locus 2 (Fig. 10-2). Suppose an individual’s genotype at these loci is Aa and Bb; that is, she is heterozygous at both loci, with alleles A and B inherited from her father and alleles a and b inherited from her mother. The two different chromosomes will line up on the metaphase plate at meiosis I in one of two combinations with equal likelihood. After recombination and chromosomal segregation are complete, there will be four possible combinations of alleles, AB, ab, Ab, and aB, in a gamete; each combination is as likely to occur as any other, a phenomenon known as independent assortment. Because AB gametes contain only her paternally derived alleles, and ab gametes only her maternally derived alleles, these gametes are designated parental. In contrast, Ab or aB gametes, each containing one paternally derived allele and one maternally derived allele, are termed nonparental gametes. On average, half (50%) of gametes will be parental (AB or ab) and 50% nonparental (Ab or aB).
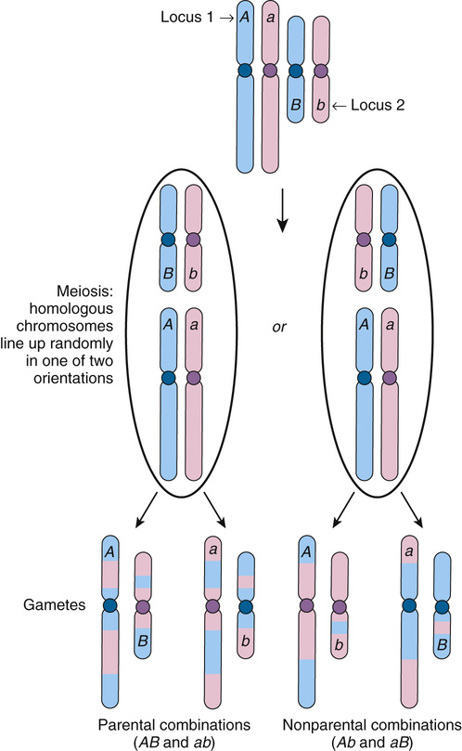
Alleles at Loci on the Same Chromosome Assort Independently If at Least One Crossover between Them Always Occurs
Now suppose that an individual is heterozygous at two loci 1 and 2, with alleles A and B paternally derived and a and b maternally derived, but the loci are on the same chromosome (Fig. 10-3). Genes that reside on the same chromosome are said to be syntenic (literally, “on the same thread”), regardless of how close together or how far apart they lie on that chromosome.
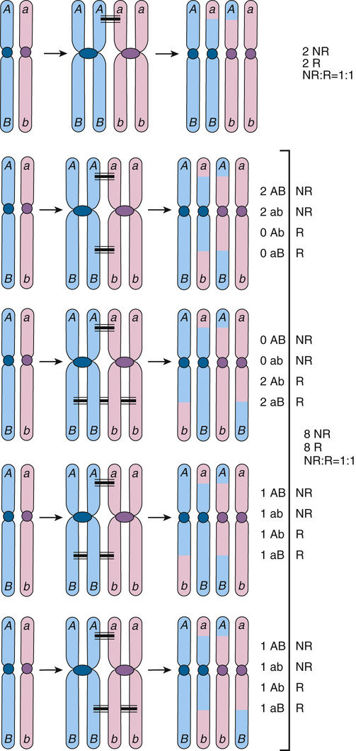
How will these alleles behave during meiosis? We know that between one and four crossovers occur between homologous chromosomes during meiosis I when there are two chromatids per homologous chromosome. If no crossing over occurs within the segment of the chromatids between the loci 1 and 2 (and ignoring whatever happens in segments outside the interval between these loci), then the chromosomes we see in the gametes will be AB and ab, which are the same as the original parental chromosomes; a parental chromosome is therefore a nonrecombinant chromosome. If crossing over occurs at least once in the segment between the loci, the resulting chromatids may be either nonrecombinant or Ab and aB, which are not the same as the parental chromosomes; such a nonparental chromosome is therefore a recombinant chromosome (shown in Fig. 10-3). One, two, or more recombinations occurring between two loci at the four-chromatid stage result in gametes that are 50% nonrecombinant (parental) and 50% recombinant (nonparental), which is precisely the same proportions one sees with independent assortment of alleles at loci on different chromosomes. Thus, if two syntenic loci are sufficiently far apart on the same chromosome to ensure that there is going to be at least one crossover between them in every meiosis, the ratio of recombinant to nonrecombinant genotypes will be, on average, 1 : 1, just as if the loci were on separate chromosomes and assorting independently.
Recombination Frequency and Map Distance
Frequency of Recombination as a Measure of Distance between Loci
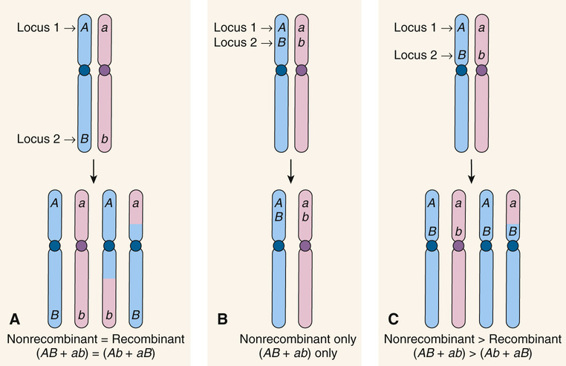
Suppose now that two loci are on the same chromosome but are either far apart, very close together, or somewhere in between (Fig. 10-4). As we just saw, when the loci are far apart (see Fig. 10-4A), at least one crossover will occur in the segment of the chromosome between loci 1 and 2, and there will be gametes of both the nonrecombinant genotypes AB and ab and recombinant genotypes Ab and aB, in equal proportions (on average) in the offspring. On the other hand, if two loci are so close together on the same chromosome that crossovers never occur between them, there will be no recombination; the nonrecombinant genotypes (parental chromosomes AB and ab in Fig. 10-4B) are transmitted together all of the time, and the frequency of the recombinant genotypes Ab and aB will be 0. In between these two extremes is the situation in which two loci are far enough apart that one recombination between the loci occurs in some meioses but not in others (see Fig. 10-4C). In this situation, we observe nonrecombinant combinations of alleles in the offspring when no crossover occurred and recombinant combinations when a recombination has occurred, but the frequency of recombinant chromosomes at the two loci will fall between 0% and 50%. The crucial point is that the closer together two loci are, the smaller the recombination frequency, and the fewer recombinant genotypes are seen in the offspring.
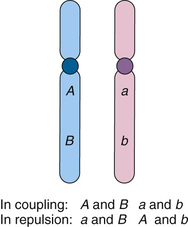
Detecting Recombination Events Requires Heterozygosity and Knowledge of Phase
Detecting the recombination events between loci requires that (1) a parent be heterozygous (informative) at both loci and (2) we know which allele at locus 1 is on the same chromosome as which allele at locus 2. In an individual who is heterozygous at two syntenic loci, one with alleles A and a, the other B and b, which allele at the first locus is on the same chromosome with which allele at the second locus defines what is referred to as the phase (Fig. 10-5). The set of alleles on the same homologue (A and B, or a and b) are said to be in coupling (or cis) and form what is referred to as a haplotype (see Chapters 7 and 8). In contrast, alleles on the different homologues (A and b, or a and B) are in repulsion (or trans) (see Fig. 10-5).