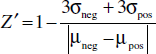Figure 1 Schematic showing the mixed dispensability present in the biosynthetic pathways of many Gram-positive cell-wall surface polymers that are assembled on a bactoprenol carrier lipid and act as virulence factors. The first two nonessential genes are typically glycosyltransferases (Gtfs), which are depicted here as light gray arrows; downstream genes shown as dark gray arrows are conditionally essential, that is, they cannot be deleted except in strains that lack one of the initiating Gtfs.
The mixed gene dispensability pattern in these bactoprenol-dependent biosynthetic pathways can be exploited in a high-throughput screen to discover inhibitors of the downstream, conditionally essential enzymes. Chemicals are tested for growth-inhibitory activities against paired strains—the wild-type strain and a mutant in which flux through the pathway has been prevented (and the downstream enzymes are no longer essential). Those molecules that stop wild-type growth, but do not inhibit the growth of the pathway-null mutant, should be selective for the conditionally essential enzymes in the pathway of interest (see Table 1). Screening against paired strains combines advantages of both in vitro and in vivo screening by limiting the targets to the conditionally essential enzymes within the desired pathway and by identifying active compounds in a cellular context. The utility of this screening strategy has been demonstrated by the discovery of an inhibitor of wall teichoic acid biosynthesis in the clinically relevant pathogen S. aureus (see Background Information and Swoboda et al., 2009, 2010).
Table 1 Concept Behind Paired Strain Screening: Small Molecules that Inhibit the Growth of the Wild-Type but not the Pathway-Null Mutant Should Target the Conditionally Essential Enzymes in the Nonessential Biosynthetic Pathway of Interest
| Wild-type | Mutanta | |
| Compound does not block any cellular process | Growth | Growth |
| Compound blocks an essential cellular process | No growth | No growth |
| Compound blocks a conditionally essential enzymeb | No growth | Growth |
aThe first enzyme in a nonessential pathway of interest is inactive.
bGrowth will be uninhibited in the mutant strain if the compound hits a conditionally essential enzyme within the deleted pathway.
The protocols outlined here allow one to design, optimize, and carry out a high-throughput screen against paired bacterial strains to find growth-inhibitory compounds that target certain enzymes within the virulence-factor pathway of interest. A simple assay using an optical density readout to measure the extent of bacterial growth is used. Calculation of Z′ values for the assay (Basic Protocol 1) allows determination of whether the chosen incubation conditions allow adequate and reproducible growth. The more robust the assay, the easier it becomes to identify screening positives with confidence. Basic Protocol 2, “Screening for growth-inhibitory compounds,” describes the steps involved in setting up a high-throughput screen. It is assumed that the screener has access to a chemical library and a pin transfer robot. Once the data have been analyzed and screening positives have been identified, Basic Protocol 3 outlines the setup of 6-point dose-response curves for secondary screening. Using this protocol, it is possible to determine the minimum inhibitory concentrations (MICs) of 48 compounds in a single 384-well plate.
STRATEGIC PLANNING
Acquiring Bacterial Strains
Mixed gene dispensability is frequently observed in biosynthetic pathways that have two things in common: (1) they make cell surface polymers and (2) they assemble them on a bactoprenol carrier. Many of these cell surface polymers function as virulence factors and share key similarities in their biosynthetic pathways: the first two genes can be knocked out with the strain remaining viable, but downstream genes cannot be knocked out unless suppressor mutations that inactivate one of the first two enzymes are present (Fig. 1). Theoretically, the screening strategy described here should be applicable to all of these biosynthetic pathways.
In order to screen paired bacterial strains for inhibitors of downstream enzymes within virulence-factor biosynthetic pathways, it is first necessary to confirm that these enzymes are conditionally essential. To do this, the first gene in the pathway is placed under an inducible promoter and then, in the absence of inducer, knockouts of the downstream genes are made. Growth curves of the resulting strains are then acquired in the absence and presence of inducer. Knockout strains of conditionally essential enzymes will be able to grow in the absence of inducer, but not in its presence (D’Elia et al., 2006; Swoboda et al., 2009). Screening is then performed against a wild-type strain and a mutant in which the first enzyme in the targeted biosynthetic pathway is inactive.
NOTE: If the goal is to screen small molecules for growth-inhibitory activities against only a single bacterial strain, the protocols outlined below can easily be adapted for this purpose.
Screening in Duplicate
The majority of high-throughput screens show a high degree of intrinsic variability and error. It is therefore recommended that these assays be run in duplicate in a second set of assay plates. This assay duplication is built into the high-throughput screening protocol described here. Acquiring duplicate data points for each library compound can reduce the rates of false positives by as much as 50%.
Biosafety Considerations
If possible, it is best to screen against bacterial strains that have a Biosafety Level 1 (BSL-1) rating. These strains can be handled on the benchtop, and no additional safety measures need to be taken. However, if screening against a BSL-2-rated pathogen, one must handle this strain in a biosafety cabinet at all times unless it is in a sealed container. When carrying out a screen using a bacterial strain that requires biocontainment, initial medium dispensing and pin transfer of compounds can be performed on the benchtop, but once the plates have been inoculated with the bacteria, they must be transported and incubated in leak-proof containers (e.g., Nalgene, cat. no. 7135). Because these containers are airtight, only ∼10 assay plates can be incubated in each one. Otherwise, because of limited oxygen, the negative controls would not grow sufficiently for optimal assay conditions. To obtain assay data when growing a BSL-2 strain, a plate reader would need to be used inside a biosafety cabinet and decontaminated before removal. Assay plates then need to be disposed of properly. These extra precautions can greatly reduce the throughput of an assay.
Selective Antibiotics
Contamination can be introduced into an assay when liquid dispensing and pin transfer steps occur on the benchtop. If the contaminating bacteria are sensitive to the positive control compound, it is almost impossible to detect their presence through standard optical density measurements. Hence, it is suggested that selective antibiotics be used throughout the screen. They should be present in the medium at all times (during both pin transfer and bacterial inoculation). Plasmid-borne selective markers have been successfully used to avoid contamination during screening (see Background Information and Swoboda et al., 2009).
BASIC PROTOCOL 1
Z′ DETERMINATION TO ASSESS SCREENING ASSAY ROBUSTNESS
For high-throughput screening, it is necessary to ensure that the readout window be sufficiently large and reproducible to enable the identification of screening positives with confidence. A standard parameter used to assess the quality and robustness of an assay is the Z′ value (Zhang et al., 1999).
This parameter requires a mean (µ) and standard deviation (σ) for both the positive and negative controls. Increasing the sample size improves the statistical significance, so half of a 384-well plate should be used for each control condition. An assay should have a Z′ of at least 0.5 to be considered robust enough for high-throughput screening, while an assay with a Z′ greater than 0.7 is considered excellent.
Materials
Sterile tryptic soy broth (TSB; 30 g/l; sterile filtered or autoclaved; BD Difco, cat. no. 211823) plus selective antibiotic, if appropriate
Plated bacterial strain(s) of interest (e.g., S. aureus)
Positive control compound that inhibits growth of bacterial strain (e.g., for S. aureus, erythromycin at 10 mg/ml in ethanol)
30°C shaking incubator
Multichannel pipettor (volume range from 20 to 100 µl) with aerosol-barrier pipet tips (VWR, cat. no. 53510-106) and sterile solvent reservoirs (VWR, cat. no. 89094-680)
384-well clear-bottom plate (Corning, cat. no. 3702)
Low-evaporation lid (Corning, cat. no. 3009)
Plate reader capable of measuring optical density at 600 nm (OD600) in a 384-well format (e.g., EnVision from PerkinElmer)
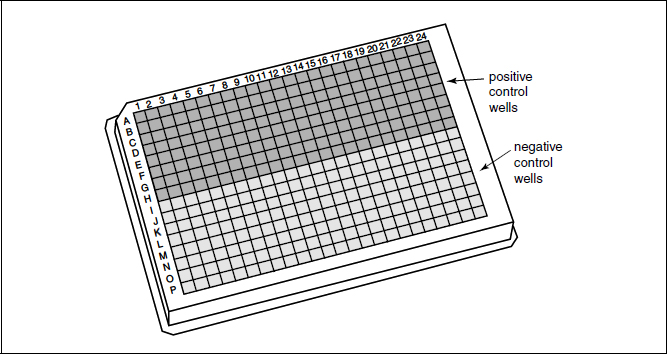
Figure 2 Schematic showing a 384-well plate layout for Z′ determination. In a growth-inhibitory assay, the positive control wells will most likely contain bacteria in medium containing erythromycin (10 µg/ml) or another appropriate antibiotic, while the negative control wells will be filled with medium and bacteria only.
BASIC PROTOCOL 2
SCREENING FOR GROWTH-INHIBITORY COMPOUNDS
To find growth-inhibitory compounds of a wild-type strain, compound library plates should be screened in duplicate (see Strategic Planning). To discover inhibitors of conditionally essential enzymes in a nonessential biosynthetic pathway, one should also screen the compound library plates in duplicate against a mutant strain that lacks the first enzyme in the pathway of interest. Those compounds that inhibit growth of the wild-type strain but not the mutant should target the conditionally essential enzymes within the desired pathway (see Table 1). This protocol is designed for one screening session of 40 compound library plates in duplicate against a single bacterial strain of interest, or for 20 compound library plates in duplicate against both a wild-type and a mutant strain (four daughter plates per library plate). For this protocol, it is assumed that columns 23 and 24 of each compound library plate are empty and thus those columns can be used for negative (column 23) and positive (column 24) controls for the assay (Fig. 3). The location of control wells can be changed as appropriate for screens using compound library plates that are formatted differently.
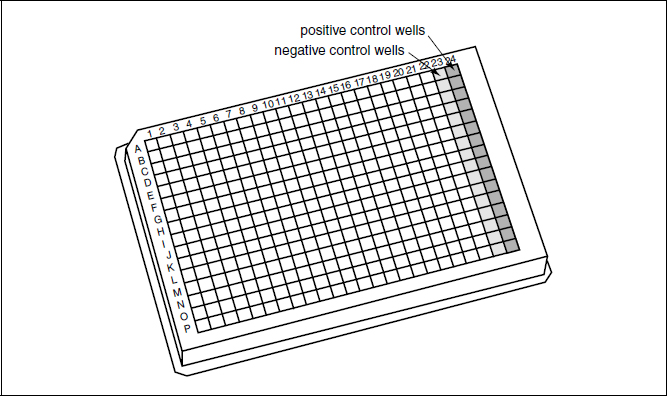
Figure 3 Schematic showing a 384-well plate layout for high-throughput screening. This format assumes that the library compounds are arrayed in columns 1 to 22, allowing for a column each of negative and positive controls. Columns 1 to 23 are filled with 40 µl of medium by the plate filler, while positive control medium (40 µl medium plus erythromycin for a 10 µg/ml final screening concentration) is added manually to column 24 using a multichannel pipettor. Following the pin transfer of small molecules from compound library plates, all wells are filled with 40 µl of medium containing the bacteria of interest.
Materials
Sterile tryptic soy broth (TSB; 30 g/l; sterile filtered or autoclaved; BD Difco, cat. no. 211823) plus selective antibiotic, if appropriate
Plated bacterial strain(s) of interest (e.g., S. aureus)
20 ml TSB medium containing positive control compound at 2× final concentration (typically erythromycin at 20 µg/ml)
Compound library plates, with compounds diluted in DMSO at 5 mg/ml or 10 mM stock concentration
1-liter flasks (sterile)
30°C shaking incubator
Multichannel pipettor (volume range from 20 to 100 µl) with aerosol-barrier pipet tips (VWR, cat. no. 53510-106) and sterile solvent reservoirs (VWR, cat. no. 89094-680)
384-well clear-bottom plates (80 per screening session; Corning, cat. no. 3702)
Microplate dispenser (Matrix WellMate or comparable liquid handler; see Rudnicki and Johnston, 2009)
Pin transfer robot (Rudnicki and Johnston, 2009)
Low-evaporation plate lids (16–20; Corning, cat. no. 3009)
Plate reader capable of measuring optical density at 600 nm (OD600) in a 384-well format (e.g., EnVision from Perkin Elmer)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree