Additional Genetic Subtypes of Acute Myeloid Leukemia, Including t(6;9), inv(3), t(1;22), Mutated NPM1, Mutated CEBPA
David Czuchlewski, MD
Key Facts
Terminology
WHO 2008 classification, adopting same philosophy as WHO 2001, revised roster of genetically defined AML subtypes
Diagnostic Checklist
Cytogenetic and molecular findings discussed in this chapter are not diagnostic evidence of AML if blasts (or blast equivalents) number < 20%
Reporting Considerations
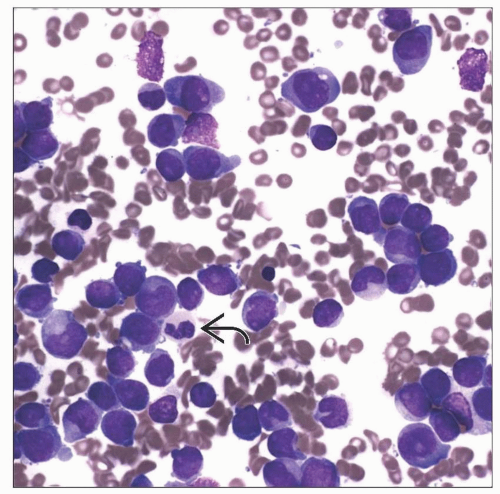
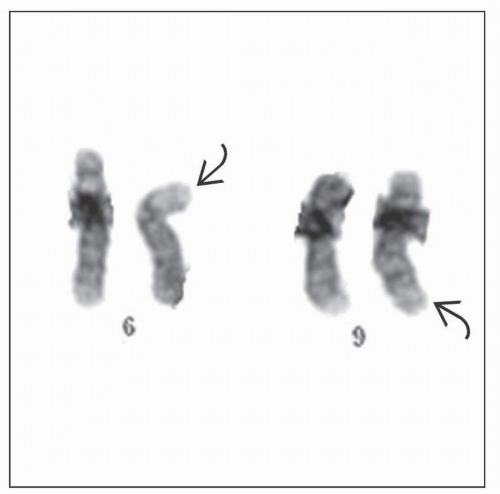
AML with t(6;9)(p23;q34)
Often presents with some degree of basophilia
AML with inv(3)(q21q26.2) or t(3;3)(q21;q26.2)
Dysplastic small, mono- and bilobated megakaryocytes are present
AML with t(1;22)(p13;q13)
Unique among entities in this chapter in being restricted to infants and children < 3 years old
Blasts are morphologically and immunophenotypically megakaryoblasts
AML with mutated NPM1
NPM1 mutation with concurrent FLT3 wild-type status is prognostically favorable, similar to that of AML with t(8;21), t(16;16), or inv(16)
This survival advantage is largely abrogated by presence of coexisting FLT3-ITD mutation
NPM1 mutation analysis should be performed in concert with FLT3-ITD mutation analysis
AML with mutated CEBPA
Prognostically favorable, similar to that of AML with t(8;21), t(16;16), or inv(16)
TERMINOLOGY
Definitions
Subclassification of acute myeloid leukemia (AML) was historically based on morphologic features
The former French-American-British classification followed this approach
WHO 2001 classification synthesized morphologic, clinical, immunophenotypic, and genetic information to define subcategories of AML
This scheme recognized 4 major types of AML characterized by recurrent genetic abnormalities
AML with t(8;21)(q22;q22)
AML with inv(16)(p13q22) or t(16;16)(p13;q22)
Acute promyelocytic leukemia [AML with t(15;17) (q22;q12) and variants]
AML with 11q23 abnormalities
WHO 2008 classification, adopting same philosophy as WHO 2001, revised roster of genetically defined AML subtypes
Instead of AML with 11q23 abnormalities, specific subset with t(9;11)(p22;q23) was recognized
Several additional types of AML defined by recurrent genetic abnormalities were added
AML with t(6;9)(p23;q34)
AML with inv(3)(q21q26.2) or t(3;3)(q21;q26.2)
AML with t(1;22)(p13;q13)
In addition, for 1st time, gene mutations (as opposed to recurrent translocations) were deemed to define provisional entities
AML with mutated NPM1
AML with mutated CEBPA
In addition, FLT3 mutations are discussed
FLT3 mutations are seen in a number of well-defined subtypes of AML
Thus, FLT3 mutations do not define specific subcategory of AML
However, FLT3 mutations can significantly affect prognosis, especially in AML with normal cytogenics
ETIOLOGY/PATHOGENESIS
AML with t(6;9)(p23;q34)
Produces fusion of DEK and NUP214
AML with inv(3)(q21q26.2) or t(3;3) (q21;q26.2)
Produces juxtaposition of RPN1 and EVI1
AML with t(1;22)(p13;q13)
Produces fusion of RBM15 and MKL1
AML with Mutated NPM1
NPM1 encodes protein nucleophosmin
Nucleophosmin appears to play several roles
Shuttles other proteins between nucleus and cytoplasm
NPM1 mutations involve small insertions in specific region of gene
Resulting frameshift causes change in nuclear localization motif in nucleophosmin
With this signal altered, protein is aberrantly concentrated in cytoplasm
Abnormal cellular localization disturbs functions of nucleophosmin and promotes transformation
AML with Mutated CEBPA
CEBPA encodes transcription factor necessary for granulocytic differentiation
CEBPA mutations abrogate this function, preventing differentiation of immature granulocytic precursors
FLT3 Mutations
FLT3 encodes tyrosine kinase receptor that promotes cellular proliferation in immature hematopoietic cells
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree