Figure 1 Structure of parainfluenza virus. Diagram of parainfluenza virus (Sendai virus, top left) and its genomic RNA (bottom). Electron micrograph of Sendai virus (top middle) and its helical nucleocapsid (top right).
Surface glycoproteins
The HN and F glycoproteins are anchored in the viral envelope by a hydrophobic transmembrane domain. Most of the protein lies external to the membrane, with a short hydrophilic cytoplasmic domain lying internal to the membrane. HN is a type-II glycoprotein which has a transmembrane domain near the N-terminus of the protein. The parainfluenza HN glycoproteins contain 4–10 sites for the addition of N-linked carbohydrate chains. Most of the parainfluenza HNs form oligomers consisting of disulphide-linked homodimers. HN is a multifunctional protein and is the virion’s main type-specific antigenic determinant. HN is responsible for binding the virus to sialic acid-containing receptors (glycolipid or glycoprotein) on cells. In addition, HN mediates enzymatic cleavage of sialic acids from cell surface receptors; this cleavage enhances the release of progeny virions from infected cells and aids their spread to uninfected target cells. The three-dimensional structure of HN revealed the protein to be a six-sheeted β-propeller, similar to influenza virus and bacterial neuraminidases (Crennell et al., 2000). The receptor-binding activity resides in the same site as the neuraminidase activity. However, some paramyxoviruses contain a 2nd receptor-binding site on HN (Zaitsev et al., 2004).
The other surface glycoprotein, F, is a type-I molecule with a transmembrane domain near the C-terminus. F mediates fusion of the virus with the host cell membrane and fusion of infected cells with adjacent cells. The F protein is synthesised in infected cells as a biologically inactive precursor protein, F0. A host cell proteolytic enzyme cleaves F0 to produce the biologically active disulphide-linked subunits F1 and F2, with F1 containing the C-terminal transmembrane domain. The sequence spanning approximately the first 25 amino acids at the N-terminus of F1 is hydrophobic and is the most highly conserved region in the F protein. These N-terminal residues are called the fusion peptide, and direct insertion of the fusion peptide into the target cell membrane is believed to play the largest role in the fusion of the virus envelope with the cell membrane. Crystal structure of the paramyxovirus F protein in two conformations, representing pre- and post-fusion states, revealed a large scale, irreversible refolding during membrane fusion (Lamb and Jardetzky, 2007).
Matrix protein
The M protein is the smallest of the major structural proteins (Mr approximately 40000) and is the most abundant protein produced in the paramyxoviruses. The parainfluenza virus M proteins contain 348–383 residues and are basic proteins. Although the M proteins do not have hydrophobic sequences long enough to span a cell membrane, the general hydrophobic and basic nature of the protein allows association with cell membranes. On the cytoplasmic side of the plasma membrane, M proteins interact with each other to form a sheet that excludes cellular membrane proteins. The M proteins are thought to associate with specific HN and F glycoproteins through their cytoplasmic domains. In addition to glycoproteins, the M proteins also recognise and associate with viral nucleocapsids. Freeze-fracture electron micrographs of virus particles show that the M protein forms a sheet between the lipid bilayer on the outside and the viral nucleocapsid on the inside. When treated with non-ionic detergents, the M protein remains bound to the nucleocapsid, although it is dissociated from the glycoproteins. The interaction between the M protein and the nucleocapsid is highly specific, and dissociation requires treatment with highly ionic reagents. Furthermore, the M protein expressed singly can trigger the budding process (Coronel et al., 1999). The expression of M protein from cDNA results in the formation of M-containing virus-like particles (VLPs), and these VLPs are released into the medium. Because of these functions, the M protein is considered to be the central organiser of viral assembly.
Nucleocapsid
The viral genome consists of a nonsegmented negative-strand RNA molecule 15000–16000 nucleotides in length. The genomic RNA is tightly associated with NP to form a helical nucleocapsid core that has a buoyant density of 1.30–1.31gcm−3 in caesium chloride (Figure 1). A viral polymerase complex that consists of P and L proteins associates with the nucleocapsid core, and together these four components compose the nucleocapsid. The Sendai virus nucleocapsid is composed of approximately 2600 NP, 300 P and 50 L protein molecules. Sequence analysis of the NP gene has shown that the N-terminal 80% of the protein is relatively well conserved among related viruses, whereas the C-terminal 20% is poorly conserved. The hypervariable C-terminus appears to be the region exposed at the surface of the nucleocapsid, and it contains the domain that binds to the M protein. The well-conserved N-terminal region contains the determinants for the NP–NP interaction that forms the helical structure and RNA-binding domain. The P protein, which was named for its highly phosphorylated nature, plays a central role in RNA synthesis, and forms homotetramers via predicted coiled-coils. Sendai virus P protein is composed of N– and C-terminal conserved domains separated by a hypervariable region. These domains interact with the NP and L proteins. The C-terminal half is essential for transcription and contains the domain required for binding to the L protein and to the nucleocapsid. The L protein has a molecular size greater than 200kDa and is the least abundant of the structural proteins. Although the precise composition of the L–P polymerase complex is unclear, it is known that the P-binding site on the L protein is located in the N-terminal half of the protein. This P–L complex is responsible for the transcription that produces messenger RNA (mRNA), which is capped at its 5′ end and polyadenylated at its 3′ end, and for the replication of the viral genome. The L proteins are thought to execute all of the catalytic steps of RNA synthesis, capping and methylation.
Accessory proteins
The accessory proteins C and V are expressed from the P gene. The V mRNA is generated by RNA editing, in which one or two G residues are inserted into the transcripts of the P gene at the editing site, except in Rubulaviruses, which produce V from intact mRNA and P from edited mRNA. Respiroviruses express C proteins from the open reading frame (ORF) that overlaps the N-terminal portion of the P gene, in the +1 frame. The accessory proteins can abrogate various facets of type I interferon (IFN) induction and signalling, and are therefore, major factors of virus pathogenicity. In addition, the C protein regulates viral genome transcription/replication through an interaction with L protein.
Genomic RNA
The complete genome sequences of many parainfluenza viruses are now known (e.g. Sendai virus, 15384 nucleotides; HPIV-3, 15462 nucleotides). The genome map of Sendai virus, one of the best-characterised parainfluenza viruses, is shown in Figure 1. The genomic RNA of all parainfluenza viruses generates six separate nonoverlapping polyadenylated mRNAs that encode NP, P, M, F, HN and L proteins. The mRNA that encodes the P protein contains several additional ORFs that encode C and V proteins. The genome consists, in 3′–5′ order, of the leader sequence, the genes for NP, P, M, F, HN and L, and the trailer sequence. The 3′ leader and 5′ trailer regions contain the viral promoters for synthesis of complementary RNA from the template RNAs. The viral polymerase complex transcribes the genome in a linear, sequential, stop–start manner guided by transcriptional start and stop signals. The Sendai virus gene start sequence consists of 10 nucleotides (3′-UCCCU/AG/C/AUUU/AC) that encode the first 10 nucleotides of the mRNA. Each gene ends with the conserved sequence 3′-C/A/UAUUCUUUUU, which encodes the 3′ end of each mRNA and encodes the poly(A) tail by reiterative copying of the U tract. The genes are separated by the trinucleotides GAA or GGG, intergenic sequences that are highly conserved among Respiroviruses. These trinucleotides, flanked on either side by the gene end and gene start sequences, are thought to participate in the signalling of transcriptional termination and initiation by the viral polymerase complex. In contrast, the intergenic sequences of Rubulaviruses are quite variable in composition and length; this variation suggests that, with Rubulaviruses, intergenic sequences do not provide cis-acting signals involved in viral transcription.
Viral Replication
Entry
Parainfluenza virus infection is initiated by viral attachment to cellular receptors. After viral HN glycoprotein binds to sialic acid-containing glycolipids or glycoproteins on the cell surface, F protein induces fusion of the viral envelope and the cellular plasma membrane. Membrane fusion occurs at a neutral pH, suggesting that the event is mediated at the cell surface. The HN protein is also involved in the fusion process. Significant membrane fusion (syncytium formation) is observed only when HN and F proteins of the same virus or of closely related viruses are expressed in the same cells (Figure 2). Thus, a virus type-specific interaction is necessary for membrane fusion induced by the F protein. In fact, it has been suggested that physical interaction between HN and F proteins of homologous viruses occurs. Experiments using chimeric and mutant HN proteins have shown that both the stalk region (close to the transmembrane domain) and the head region are responsible for promoting fusion activity (Bousse et al., 1994). Solution of the atomic structure of HN and a subsequent mutagenesis study provided insight into the role of HN in membrane fusion (Takimoto et al., 2002). The fusion promotion domain is located at the hydrophobic surface of the HN protein, which includes the area where the structure changes upon binding to the receptor. A current model for the fusion process is that a structural change in HN induced by receptor-binding triggers a conformational change in F that exposes the fusion peptide. The fusion peptide is then inserted into the cell membrane to mediate fusion with the viral envelope. In this way, F and HN operate together as a molecular scaffold to expose and direct the fusion peptide to the target membrane (Baker et al., 1999; Takimoto et al., 2002).
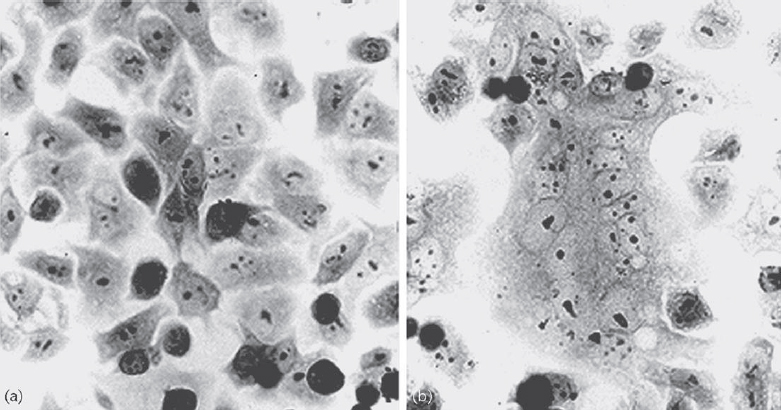
Figure 2 Syncytium formation induced by haemagglutinin-neuraminidase (HN) and fusion (F) proteins. Membrane fusion occurs when HN and F proteins of the homologous viruses (b) but not heterologous viruses (a) are expressed; this finding indicates that specific interaction between HN and F is required for membrane fusion (Bousse et al., 1994).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


