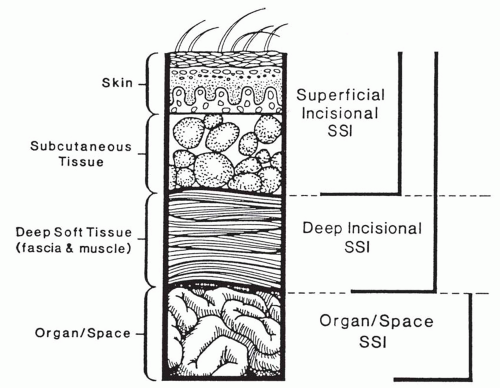
Surgical Site Classification
The risk of developing an SSI is affected by the degree of microbial contamination of the operative site. A widely accepted system of classifying operative site contamination was developed by the National Research Council for its cooperative study of the effects of ultraviolet irradiation of operating rooms on SSIs
(36), with the least contamination in clean sites and the most in dirty-infected sites. This classification scheme, in a modified form, is as follows:
Clean sites (wounds): These are surgical sites in which no inflammation is encountered and the respiratory, alimentary, genital, and urinary tracts are not entered. In addition, clean wounds are primarily closed and, if necessary, drained with closed drainage. Surgical sites for operations that follow nonpenetrating (blunt) trauma should be included in this category if they meet these criteria.
Clean-contaminated sites (wounds): These are operative sites in which the respiratory, alimentary, genital, or urinary tract is entered under controlled conditions and without unusual contamination. Specifically, operations involving the biliary tract, appendix, vagina, and oropharynx are included in this category, provided no evidence of infection or major break in technique is encountered.
Contaminated sites (wounds): These include open, fresh accidental wounds or operations with major breaks in sterile technique or gross spillage from the gastrointestinal tract. Surgical sites through which there is entry into the genitourinary tract with infected urine or biliary tract with infected bile, and surgical sites in which acute, nonpurulent inflammation is encountered, fall into this category.
Dirty and infected sites (wounds): These include old traumatic wounds with retained devitalized tissue, foreign bodies, or fecal contamination. Surgical sites where a perforated viscus or pus is encountered during the operation fall into this category.
Early studies showed that this surgical site (wound) classification scheme did predict the risk of subsequent SSIs. In Cruse and Foord’s
(37) study, surgery involving clean, clean-contaminated, contaminated, and dirty surgical sites had infection rates of 1.5%, 7.7%, 15.2%, and 40%, respectively. The SSI rates in the National Research Council cooperative study were 3.3% for refined clean sites, 7.4% for other clean sites, 16.4% for contaminated sites, and 28.6% for dirty sites
(36).
The correlation of site (wound) class to the risk of SSIs would suggest that intraoperative site contamination should also be linked to the risk of subsequent infections. However, conflicting results were obtained when the microbiology of intraoperative site contamination was examined and attempts were made to correlate microorganisms isolated intraoperatively with pathogens responsible for the SSIs. Barlett et al.
(38) isolated bacteria from 43 of 91 (47%) intraoperative surgical site irrigation cultures. However, they found no significant difference in the rate of subsequent SSIs between those patients with and those without positive cultures. Further, there was no relationship between the concentration of bacteria in the sites and the subsequent development of infection. A more recent prospective study of neurosurgical patients found no association between total colony-forming unit (CFU) counts of skin flora, either before or after skin preparation, at the operative site and SSIs
(39). Therefore, it is clear that degree of microbial contamination is only one risk factor for development of SSIs.
Sources for Pathogens Causing Surgical Site Infections
Pathogens that cause SSIs are predominately acquired endogenously from the patient’s own flora or potentially from exogenous contact with operating room personnel or the environment. It is believed that, within 24 hours of an operative procedure, most surgical sites are sufficiently sealed, unless the site was closed secondarily or involved drain placement, making the surgical site resistant to inoculation and infection. Thus, most pathogens, whether endogenously or exogenously acquired, are believed to be implanted at the time of surgery
(40). Theoretically, the operative site can be seeded postoperatively by the hematogenous or lymphatic route or by direct inoculation of the closed operative site, but such mechanisms of acquisition are thought to occur infrequently
(40). Ehrenkranz and Pfaff
(41), however, described a cluster of sternal infections occurring postoperatively that were preceded by infections caused by the same microorganisms at remote sites (pneumonias and bacteremias). In the outbreak of
Legionella sternal infections reported by Lowry et al.
(34), patients were not exposed to contaminated tap water containing
Legionella during bathing and dressing changes until well after cardiac surgery. Thus, there is evidence to suggest that inoculation (and infection) may occasionally occur postoperatively. Nonetheless, the period of greatest risk for infection remains the time between opening and closing the operative site.
Endogenous Sources of Pathogens The patient’s own flora at or contiguous to the site of operation accounts for the majority of SSIs
(42). S. aureus and coagulase-negative staphylococci, the first and second most frequent causative microorganisms, are residents of skin and mucous membranes, and presumably they are directly inoculated
into the operative site during incision or subsequent manipulations. Between 2006 and 2007, 44% of all SSIs reported to the NHSN were either due to coagulase-negative
Staphylococcus or
S. aureus; over 56% of SSIs were due to gram-positive microorganisms and yeast, common skin commensals (
12). Unsurprisingly, colonization of the nares and skin with
S. aureus is a risk factor for developing SSI due to
S. aureus, and may quadruple the odds of a
S. aureus SSI compared to those who are not colonized (
43). Recently, in a double-blind randomized trial, rapid identification of
S. aureus colonization by PCR, and subsequent decolonization of the skin and nares of colonized individuals with chlorhexidine showed a 60% reduction in cardiac surgery SSIs due to
S. aureus (
44). However, in order to prevent one
S. aureus SSI, the number needed to screen and treat was 250 and 23, respectively, making screening and decolonization not cost-effective.
Skin antisepsis during preparation of the operative site for surgery is routinely performed and reduces the surface population of all skin microorganisms, therefore reducing risk of SSIs. Darouiche et al. (
45) recently published a prospective randomized study of patients undergoing clean-contaminated surgery (70% abdominal, 30% nonabdominal), which demonstrated a >40% reduction in total SSIs among patients randomized to preoperative chlorhexidine-alcohol skin preparation compared to providine-iodine scrub. This decrease was due to a significant decline in incidence of superficial and deep incision infections caused by gram-positive bacteria and
Candida, demonstrating the importance of skin flora on incisional SSI pathogenesis, even among clean-contaminated surgeries.
However, if the skin became heavily colonized—for example, as a result of dermatitis—resident flora may persist and be carried into the operative site. In addition, even optimal skin antisepsis may not be able to eradicate all skin bacteria, as up to 20% of these bacteria live beneath the skin’s surface along the hair follicles and sebaceous glands
(40).
During nonclean surgery, besides the significant role of skin flora that can contaminate the incision, normal flora of the gastrointestinal, respiratory, genital, and urinary tracts can directly contaminate the operative site when these tracts are opened or when injury has occurred to one of these tracts prior to surgery.
The patient’s endogenous flora at distant sites may also be a source of SSI. Wiley and Ha’eri
(46) noted that human albumin microspheres (HAMs) were like human skin squames and could be used as tracer particles. When they applied HAM to the patient’s skin outside the area of the incision, they demonstrated that the tracer particles could be easily recovered from the operative site (in 40 of 40 orthopedic operations), suggesting that surface microflora can migrate from distant sites and gain entrance to the operative site despite distance and the use of cloth and adhesive drapes as barriers. Finally, microorganisms causing infections at remote sites may gain access to operative sites by hematogenous or lymphogenous seeding, which is most commonly associated with bacteremia after implantation of prosthetic material (
47). Untreated urinary tract, skin, and respiratory tract infections have also been associated with an increase in the rate of SSIs
(48,
49).
Exogenous Sources of Pathogens Personnel The hands and nails of the operative team harbor microorganisms that can contaminate the surgical site by direct inoculation during the operative procedure
(50,
51,
52). This has led to the use of surgical gloves as a barrier to the transfer of microorganisms and to the surgical hand scrub to reduce the microbial population on the skin of the hands. Initially introduced as a way of protecting operating room personnel against dermatitis from Listerian antisepsis, surgical gloving has became a standard of practice as a method to prevent the passage of microorganisms from the surgeon’s hands to the patient’s surgical site. Whether surgical gloves are an effective barrier has been questioned, since studies have demonstrated that glove perforations occur frequently; this occurs in up to a third or more of operations
(37,
52). Nonetheless, with appropriate preoperative scrubbing to reduce the burden of microorganisms on the surgeon’s hands, there is no evidence that such perforations of surgical gloves are of any clinical significance. Dodds et al.
(53) found no difference in the rate of SSIs among 100 hernia repairs that were or were not associated with glove perforations.
However, despite standard hand hygiene and gloving, outbreaks due to artificial nails have been reported, due to sequestered microorganisms trapped between the natural and artificial nail
(54).
In addition to the hands, other body sites in the operative team may be sources for exogenous contamination of the operative site. The hair and scalp of hospital staff (as well as of patients themselves), nares and oropharynges have been shown to harbor potentially pathogenic bacteria, including
S. aureus and gram-negative bacteria
(55). Despite those observations, however, only a few outbreaks of SSIs have been traced to the hair/scalp or nasopharynx of the operative team
(50,
56). However, outbreaks of group A
Streptococcus SSI have been traced to anal or vaginal carriage by operating room personnel
(57,
58,
59,
60).
Environment The microorganisms that are isolated from the operating room environment are usually considered nonpathogens or commensals that are rarely associated with infections (
61). Atypical mycobacteria are ubiquitous and can be recovered from hospital dust but are rarely incriminated in SSIs. In the clusters of infections due to Mycobacterium fortuitum and M. chelonae that followed valve replacement surgery and augmentation mammoplasty (
27,
28,
29), it was bone wax or gentian violet marking solution that was incriminated rather than the general operating room environment. Spores of Clostridium perfringens have been isolated from the ventilation system and floors of operating rooms (
62), but when investigators looked for potential sources for these microorganisms that cause devastating SSIs, they concluded that
C. perfringens was either endogenously acquired from the patient’s own gastrointestinal flora (
63) or acquired from contaminated surgical instruments that had been inadequately sterilized between cases (
64).
In those rare instances when inanimate sources in the operating room have been incriminated, the sources have been contaminated solutions, antiseptics, or dressings. Contaminated elastic dressings have been implicated
in SSIs caused by
Rhizopus (24,
25,
65) and
C. perfringens (66). Contaminated solutions have been the source for SSIs caused by
P. aeruginosa, P. multivorans, and
Serratia marcescens (67,
68,
69).
It is currently standard practice to wet mop the floor of the operating room with a disinfectant between cases. Coupled with a more thorough wet vacuuming of the rooms and corridors at night, this routine is believed to provide a sufficiently clean environment that minimizes the risk of the operating room environmental surfaces and floors as a source of infection.
Air The role of the operating room air as a source of infection and the need for special ventilation systems in the operating room have long been subjects of debate. The largest source of airborne microbial contamination is the staff in the operating room (
61,
62). It is presumed that microorganisms become airborne as a result of conversation, which creates droplet nuclei from the respiratory tract, or as a result of shedding from hair or exposed skin. Tracer particle studies using HAMs suggest that airborne microorganisms from the respiratory tract or the head and neck area of operating room personnel can settle on the operative site (
46,
70,
71). Despite this possibility, there is little evidence that the airborne route of transmission contributes significantly to SSIs. Evidence that SSI resulting from airborne contamination occurs at all is based on outbreaks of group A b-hemolytic streptococcal infections that have been reported in the literature (
57,
58,
59,
60). In these outbreaks, the evidence for airborne transmission was as follows. First, streptococci with the same serotype as the isolates from infected surgical sites were isolated from sites of colonization (anal, vaginal, or pharyngeal) in operating room personnel. Second, the sites of carriage (anal or vaginal) had no possibility of direct contact with the operative site. Moreover, some of these carriers were ancillary personnel who, while they were in the same room, did not work directly in the operative field. Finally, when settling plates were used during these investigations, the epidemic microorganism could be recovered from the air of a room during exercise by the carrier.
Additional evidence for the role of airborne transmission comes from studies on the use of laminar flow air systems and ultraviolet irradiation to provide ultraclean air. Early studies appeared to show a reduction in SSIs when special air-handling systems were used to reduce airborne microbial contamination
(72,
73,
74,
75). However, many of these studies were flawed, because they were not comparative, had inadequate sample sizes, were not randomized or blinded, or included other interventions that could affect the rate of SSIs. A well-designed multicenter European study compared infection rates among total hip and knee replacement procedures that were performed in rooms with ultraclean air provided by special ventilation systems, antimicrobial prophylaxis alone, or ultraclean air plus antimicrobial prophylaxis (
76). In rooms with ultraclean air, the frequency of SSIs decreased from 3.6% to 1.6%; however, when antimicrobial prophylaxis alone was used, the rates dropped from 3.4% to 0.8%. The combination of interventions decreased rates from 3.4% to 0.7%. These results helped demonstrate antimicrobial prophylaxis to be more beneficial in prevention of SSIs than ultraclean air, with no additional benefit of ultraclean air when antibiotics were used.