KEY POINTS
Biological sciences developed drastically in last 60 years after the uncovering of DNA structure by Watson and Crick
The completion of the human genome sequence in 2003 represents a great milestone in modern science.
The technology emerging from molecular and cellular biology has revolutionized the understanding of disease and will radically transform the practice of surgery.
The use of genetically modified mouse models and cell lines using gene therapy and RNA interference therapy has greatly contributed to the understanding of the molecular basis for human diseases and targeted therapies.
The sequencing of each individual’s genome has the potential to improve the predication, prevention, and targeted treatment of disease, resulting in personalized medicine and surgery.
The use of functional genomics and modern molecular analyses will facilitate the discovery of actionable genes to guide choice of care.
OVERVIEW OF MOLECULAR CELL BIOLOGY
The beginning of modern medicine can be traced back to centuries ago when physicians and scientists began studying human anatomy from cadavers in morgues and animal physiology following hunting expeditions. Gradually, from the study of animals and plants in greater detail and the discovery of microbes, scientific principles governing life lead to the emergence of the biological sciences. As biological science developed and expanded, scientists and physicians began to utilize the principles of biological sciences to solve challenges of human diseases while continuing to explore the fundamentals of life in greater detail. With ever-evolving state-of-the-art scientific tools, our understanding of how cells, tissues, organs, and entire organisms function, down to the level of molecular and subatomic structure, has resulted in modern biology with an enormous impact on modern healthcare and the discovery of amazing treatments for disease at an exponential pace. Significant progress has been made in molecular studies of organ development, cell signaling, and gene regulation. The advent of recombinant DNA technology, polymerase chain reaction (PCR) techniques, and next-generation genomic sequencing, which resulted in the sequencing of the human genome, holds the potential to have a transformational influence on healthcare and society this century by not only broadening our understanding of the pathophysiology of disease, but also by bringing about necessary changes in personalized medicine.
Today’s practicing surgeons are becoming increasingly aware that many modern surgical procedures rely on the information gained through molecular research (i.e., personalized surgery). Genomic information, such as deleterious BRCA and RET proto-oncogene mutations, is being used to help direct prophylactic procedures to remove potentially harmful tissues before they do damage to patients. Molecular engineering has led to cancer-specific gene therapy that could serve in the near future as a more effective adjunct to surgical debulking of tumors than radiation or chemotherapy, so surgeons will benefit from a clear introduction to how basic biochemical and biological principles relate to the developing area of molecular biology. This chapter reviews the current information on modern molecular biology for the surgical community.
The modern era of molecular biology, which has been mainly concerned with how genes govern cell activity, began in 1953 when James D. Watson and Francis H. C. Crick made one of the greatest scientific discoveries by deducing the double-helical structure of deoxyribonucleic acid (DNA).1,2 The year 2003 marked the 50th anniversary of this great discovery. In the same year, the Human Genome Project completed with sequencing approximately 20,000 to 25,000 genes and 3 billion base pairs in human DNA.3 Before 1953, one of the most mysterious aspects of biology was how genetic material was precisely duplicated from one generation to the next. Although DNA had been implicated as genetic material, it was the base-paired structure of DNA that provided a logical interpretation of how a double helix could “unzip” to make copies of itself. This DNA synthesis, termed replication, immediately gave rise to the notion that a template was involved in the transfer of information between generations, and thus confirmed the suspicion that DNA carried an organism’s hereditary information.
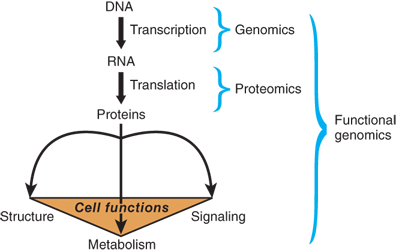
Within cells, DNA is packed tightly into chromosomes. One important feature of DNA as genetic material is its ability to encode important information for all of a cell’s functions (Fig. 15-1). Based on the principles of base complementarity, scientists also discovered how information in DNA is accurately transferred into the protein structure. DNA serves as a template for RNA synthesis, termed transcription, including messenger RNA (mRNA, or the protein-encoding RNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). mRNA carries the information from DNA to make proteins, termed translation, with the assistance of rRNA and tRNA. Each of these steps is precisely controlled in such a way that genes are properly expressed in each cell at a specific time and location. In recent years, new classes of noncoding RNAs (ncRNA), for example, microRNA (or miRNA), Piwi-interacting RNA (or piRNA), and long intergenic noncoding RNA (or lincRNA), have been identified. Although the number of ncRNAs encoded in the human genome is unknown and a lot of ncRNAs have not been validated for their functions, ncRNAs have been associated to regulate gene expression through posttranscriptional gene regulation such as mRNA degradation or epigenetic regulation such as chromatin structure modification and DNA methylation induction.4 Consequently, the differential gene activity in a cell determines its actions, properties, and functions.
Figure 15-1.
The flow of genetic information from DNA to protein to cell functions. The process of transmission of genetic information from DNA to RNA is called transcription, and the process of transmission from RNA to protein is called translation. Proteins are the essential controlling components for cell structure, cell signaling, and metabolism. Genomics and proteomics are the study of the genetic composition of a living organism at the DNA and protein level, respectively. The study of the relationship between genes and their cellular functions is called functional genomics.
Rapid advances in molecular and cellular biology over the past half century have revolutionized the understanding of disease and will radically transform the practice of surgery. In the future, molecular techniques will be increasingly applied to surgical disease and will lead to new strategies for the selection and implementation of operative therapy. Surgeons should be familiar with the fundamental principles of molecular and cellular biology so that emerging scientific breakthroughs can be translated into improved care of the surgical patient.
The greatest advances in the field of molecular biology have been in the areas of analysis and manipulation of DNA.1 Since Watson and Crick’s discovery of DNA structure, an intensive effort has been made to unlock the deepest biologic secrets of DNA. Among the avalanche of technical advances, one discovery in particular has drastically changed the world of molecular biology: the uncovering of the enzymatic and microbiologic techniques that produce recombinant DNA. Recombinant DNA technology involves the enzymatic manipulation of DNA and, subsequently, the cloning of DNA. DNA molecules are cloned for a variety of purposes including safeguarding DNA samples, facilitating sequencing, generating probes, and expressing recombinant proteins in one or more host organisms. DNA can be produced by a number of means, including restricted digestion of an existing vector, PCR, and cDNA synthesis. As DNA cloning techniques have developed over the last quarter century, researchers have moved from studying DNA to studying the functions of proteins, and from cell and animal models to molecular therapies in humans. Expression of recombinant proteins provides a method for analyzing gene regulation, structure, and function. In recent years, the uses for recombinant proteins have expanded to include a variety of new applications, including gene therapy and biopharmaceuticals. The basic molecular approaches for modern surgical research include DNA cloning, cell manipulation, disease modeling in animals, and clinical trials in human patients.
FUNDAMENTALS OF MOLECULAR AND CELL BIOLOGY
DNA forms a right-handed, double-helical structure that is composed of two antiparallel strands of unbranched polymeric deoxyribonucleotides linked by phosphodiester bonds between the 5′ carbon of one deoxyribose moiety to the 3′ carbon of the next (Fig. 15-2). DNA is composed of four types of deoxyribonucleotides: adenine (A), cytosine (C), guanine (G), and thymine (T). The nucleotides are joined together by phosphodiester bonds. In the double-helical structure deduced by Watson and Crick, the two strands of DNA are complementary to each other. Because of size, shape, and chemical composition, A always pairs with T, and C with G, through the formation of hydrogen bonds between complementary bases that stabilize the double helix.
Figure 15-2.
Schematic representation of a DNA molecule forming a double helix. DNA is made of four types of nucleotides, which are linked covalently into a DNA strand. A DNA molecule is composed of two DNA strands held together by hydrogen bonds between the pair bases. The arrowheads at the ends of the DNA strands indicate the polarities of the two strands, which run antiparallel to each other in the DNA molecule. The diagram at the bottom left of the figure shows the DNA molecule straightened out. In reality, the DNA molecule is twisted into a double helix, of which each turn of DNA is made up of 10.4 nucleotide pairs, as shown on the right. (Republished with permission of Garland Publishing, Inc. from Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell, 5th ed. New York: Garland Science; 2008. Permission conveyed through Copyright Clearance Center, Inc.)
Recognition of the hereditary transmission of genetic information is attributed to the Austrian monk, Gregor Mendel. His seminal work, ignored upon publication until its rediscovery in 1900, established the laws of segregation and of independent assortment. These two principles established the existence of paired elementary units of heredity and defined the statistical laws that govern them.5 DNA was isolated in 1869, and a number of important observations of the inherited basis of certain diseases were made in the early part of the twentieth century. Although today it appears easy to understand how DNA replicates, before the 1950s, the idea of DNA as the primary genetic material was not appreciated. The modern era of molecular biology began in 1944 with the demonstration that DNA was the substance that carried genetic information. The first experimental evidence that DNA was genetic material came from simple transformation experiments conducted in the 1940s using Streptococcus pneumoniae. One strain of the bacteria could be converted into another by incubating it with DNA from the other, just as the treatment of the DNA with deoxyribonuclease would inactivate the transforming activity of the DNA. Similarly, in the early 1950s, before the discovery of the double-helical structure of DNA, the entry of viral DNA and not the protein into the host bacterium was believed to be necessary to initiate infection by the bacterial virus or bacteriophage. Key historical events concerning genetics are outlined in Table 15-1.
YEAR | INVESTIGATOR | EVENT |
|---|---|---|
1865 | Mendel | Laws of genetics established |
1869 | Miescher | DNA isolated |
1905 | Garrod | Human inborn errors of metabolism |
1913 | Sturtevant | Linear map of genes |
1927 | Muller | X-rays cause inheritable genetic damage |
1928 | Griffith | Transformation discovered |
1941 | Beadle and Tatum | “One gene, one enzyme” concept |
1944 | Avery, MacLeod, McCarty | DNA as material of heredity |
1950 | McKlintock | Existence of transposons confirmed |
1953 | Watson and Crick | Double-helical structure of DNA |
1957 | Benzer and Kornberg | Recombination and DNA polymerase |
1966 | Nirenberg, Khorana, Holley | Genetic code determined |
1970 | Temin and Baltimore | Reverse transcriptase |
1972 | Cohen, Boyer, Berg | Recombinant DNA technology |
1975 | Southern | Transfer of DNA fragments from sizing gel to nitrocellulose (Southern blot) |
1977 | Sanger, Maxim, Gilbert | DNA sequencing methods |
1982 | — | GenBank database established |
1985 | Mullis | Polymerase chain reaction |
1986 | — | Automated DNA sequencing |
1989 | Collins | Cystic fibrosis gene identified by positional cloning and linkage analysis |
1990 | — | Human Genome Project initiated |
1997 | Roslin Institute | Mammalian cloning (Dolly) |
2001 | IHGSC and Celera Genomics | Draft versions of human genome sequence published |
2003 | — | Human Genome Project completed |
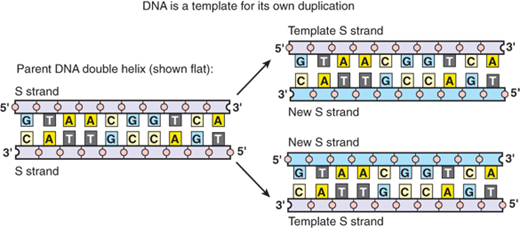
For cells to pass on the genetic material (DNA) to each progeny, the amount of DNA must be doubled. Watson and Crick recognized that the complementary base-pair structure of DNA implied the existence of a template-like mechanism for the copying of genetic material.1 The transfer of DNA material from the mother cell to daughter cells takes place during somatic cell division (also called mitosis). Before a cell divides, DNA must be precisely duplicated. During replication, the two strands of DNA separate, and each strand creates a new complementary strand by precise base-pair matching (Fig. 15-3). The two, new, double-stranded DNAs carry the same genetic information, which can then be passed on to two daughter cells. Proofreading mechanisms ensure that the replication process occurs in a highly accurate manner. The fidelity of DNA replication is absolutely crucial to maintaining the integrity of the genome from generation to generation. However, mistakes can still occur during this process, resulting in mutations, which may lead to a change of the DNA’s encoded protein and, consequently, a change of the cell’s behavior. The reliable dependence of many features of modern organisms on subtle changes in genome is linked to Mendelian inheritance and also contributes to the processes of Darwinian evolution. In addition, massive changes, so-called genetic instability, can occur in the genome of somatic cells such as cancer cells.
Figure 15-3.
DNA replication. As the nucleotide A only pairs with T, and G with C, each strand of DNA can determine the nucleotide sequence in its complementary strand. In this way, double-helical DNA can be copied precisely. (Republished with permission of Garland Publishing, Inc. from Alberts B, Johnson A, Lewis J,et al. Molecular Biology of the Cell, 5th ed. New York: Garland Science; 2008. Permission conveyed through Copyright Clearance Center, Inc.)
Living cells have the necessary machinery to enzymatically transcribe DNA into RNA and translate the mRNA into protein. This machinery accomplishes the two major steps required for gene expression in all organisms: transcription and translation (Fig. 15-4). However, gene regulation is far more complex, particularly in eukaryotic organisms. For example, many gene transcripts must be spliced to remove the intervening sequences. The sequences that are spliced off are called introns, which appear to be useless, but in fact may carry some regulatory information. The sequences that are joined together, and are eventually translated into protein, are called exons. Additional regulation of gene expression includes modification of mRNA, control of mRNA stability, and its nuclear export into cytoplasm (where it is assembled into ribosomes for translation). After mRNA is translated into protein, the levels and functions of the proteins can be further regulated posttranslationally. However, the following sections will mainly focus on gene regulation at transcriptional and translational levels.
Figure 15-4.
Four major steps in the control of eukaryotic gene expression. Transcriptional and posttranscriptional control determine the level of messenger RNA (mRNA) that is available to make a protein, while translational and posttranslational control determine the final outcome of functional proteins. Note that posttranscriptional and posttranslational controls consist of several steps.
Transcription is the enzymatic process of RNA synthesis from DNA.6 In bacteria, a single RNA polymerase carries out all RNA synthesis, including that of mRNA, rRNA, and tRNA. Transcription often is coupled with translation in such a way that an mRNA molecule is completely accessible to ribosomes, and bacterial protein synthesis begins on an mRNA molecule even while it is still being synthesized. Therefore, a discussion of gene regulation with a look at the simpler prokaryotic system precedes that of the more complex transcription and posttranscriptional regulation of eukaryotic genes.
Initiation of transcription in prokaryotes begins with the recognition of DNA sequences by RNA polymerase. First, the bacterial RNA polymerase catalyzes RNA synthesis through loose binding to any region in the double-stranded DNA and then through specific binding to the promoter region with the assistance of accessory proteins called σ factors (sigma factors). A promoter region is the DNA region upstream of the transcription initiation site. RNA polymerase binds tightly at the promoter sites and causes the double-stranded DNA structure to unwind. Consequently, few nucleotides can be base-paired with the DNA template to begin transcription. Once transcription begins, the σ factor is released. The growing RNA chain may begin to peel off as the chain elongates. This occurs in such a way that there are always about 10 to 12 nucleotides of the growing RNA chains that are base-paired with the DNA template.
The bacterial promoter contains a region of about 40 bases that include two conserved elements called –35 region and –10 region. The numbering system begins at the initiation site, which is designated +1 position, and counts backward (in negative numbers) on the promoter and forward on the transcribed region. Although both regions on different promoters are not the same sequences, they are fairly conserved and very similar. This conservation provides the accurate and rapid initiation of transcription for most bacterial genes. It is also common in bacteria that one promoter serves to transcribe a series of clustered genes, called an operon. A single transcribed mRNA contains a series of coding regions, each of which is later independently translated. In this way, the protein products are synthesized in a coordinated manner. Most of the time, these proteins are involved in the same metabolic pathway, thus demonstrating that the control by one operon is an efficient system. After initiation of transcription, the polymerase moves along the DNA to elongate the chain of RNA, although at a certain point, it will stop. Each step of RNA synthesis, including initiation, elongation, and termination, will require the integral functions of RNA polymerase as well as the interactions of the polymerase with regulatory proteins.
Transcription mechanisms in eukaryotes differ from those in prokaryotes. The unique features of eukaryotic transcription are as follows: (a) Three separate RNA polymerases are involved in eukaryotes: RNA polymerase I transcribes the precursor of 5.8S, 18S, and 28S rRNAs; RNA polymerase II synthesizes the precursors of mRNA as well as microRNA; and RNA polymerase III makes tRNAs and 5S rRNAs. (b) In eukaryotes, the initial transcript is often the precursor to final mRNAs, tRNAs, and rRNAs. The precursor is then modified and/or processed into its final functional form. RNA splicing is one type of processing to remove the noncoding introns (the region between coding exons) on an mRNA. (c) In contrast to bacterial DNA, eukaryotic DNA often is packaged with histone and nonhistone proteins into chromatins. Transcription will only occur when the chromatin structure changes in such a way that DNA is accessible to the polymerase. (d) RNA is made in the nucleus and transported into cytoplasm, where translation occurs. Therefore, unlike bacteria, eukaryotes undergo uncoupled transcription and translation.
Eukaryotic gene transcription also involves the recognition and binding of RNA polymerase to the promoter DNA. However, the interaction between the polymerase and DNA is far more complex in eukaryotes than in prokaryotes. Because the majority of studies have been focused on the regulation and functions of proteins, this chapter primarily focuses on how protein-encoding mRNA is made by RNA polymerase II.
DNA directs the synthesis of RNA; RNA in turn directs the synthesis of proteins. Proteins are variable-length polypeptide polymers composed of various combinations of 20 different amino acids and are the working molecules of the cell. The process of decoding information on mRNA to synthesize proteins is called translation (see Fig. 15-1). Translation takes place in ribosomes composed of rRNA and ribosomal proteins. The numerous discoveries made during the 1950s made it easy to understand how DNA replication and transcription involve base-pairing between DNA and DNA or DNA and RNA. However, at that time, it was still impossible to comprehend how mRNA transfers the information to the protein-synthesizing machinery. The genetic information on mRNA is composed of arranged sequences of four bases that are transferred to the linear arrangement of 20 amino acids on a protein. Amino acids are characterized by a central carbon unit linked to four side chains: an amino group (–NH2), a carboxy group (–COOH), a hydrogen, and a variable (–R) group. The amino acid chain is assembled via peptide bonds between the amino group of one amino acid and the carboxy group of the next. Because of this decoding, the information carried on mRNA relies on tRNA. Translation involves all three RNAs. The precise transfer of information from mRNA to protein is governed by genetic code, the set of rules by which codons are translated into an amino acid (Table 15-2). A codon, a triplet of three bases, codes for one amino acid. In this case, random combinations of the four bases form 4 × 4 × 4, or 64 codes. Because 64 codes are more than enough for 20 amino acids, most amino acids are coded by more than one codon. The start codon is AUG, which also corresponds to methionine; therefore, almost all proteins begin with this amino acid. The sequence of nucleotide triplets that follows the start codon signal is termed the reading frame. The codons on mRNA are sequentially recognized by tRNA adaptor proteins. Specific enzymes termed aminoacyl-tRNA synthetases link a specific amino acid to a specific tRNA. The translation of mRNA to protein requires the ribosomal complex to move stepwise along the mRNA until the initiator methionine sequence is identified. In concert with various protein initiator factors, the methionyl-tRNA is positioned on the mRNA and protein synthesis begins. Each new amino acid is added sequentially by the appropriate tRNA in conjunction with proteins called elongation factors. Protein synthesis proceeds in the amino-to-carboxy-terminus direction.
| SECOND BASE IN CODON | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | C | A | G | ||||||||||||
First Base in Codon | U | UUU | Phe | [F] | UCU | Ser | [S] | UAU | Tyr | [Y] | UGU | Cys | [C] | U | Third Base in Codon |
UUC | Phe | [F] | UCC | Ser | [S] | UAC | Tyr | [Y] | UGC | Cys | [C] | C | |||
UUA | Leu | [L] | UCA | Ser | [S] | UAA | STOP | — | UGA | STOP | — | A | |||
UUG | Leu | [L] | UCG | Ser | [S] | UAG | STOP | — | UGG | Trp | [W] | G | |||
C | CUU | Leu | [L] | CCU | Pro | [P] | CAU | His | [H] | CGU | Arg | [R] | U | ||
CUC | Leu | [L] | CCC | Pro | [P] | CAC | His | [H] | CGC | Arg | [R] | C | |||
CUA | Leu | [L] | CCA | Pro | [P] | CAA | Gln | [Q] | CGA | Arg | [R] | A | |||
CUG | Leu | [L] | CCG | Pro | [P] | CAG | Gln | [Q] | CGG | Arg | [R] | G | |||
A | AUU | Ile | [I] | ACU | Thr | [T] | AAU | Asn | [N] | AGU | Ser | [S] | U | ||
AUC | Ile | [I] | ACC | Thr | [T] | AAC | Asn | [N] | AGC | Ser | [S] | C | |||
AUA | Ile | [I] | ACA | Thr | [T] | AAA | Lys | [K] | AGA | Arg | [R] | A | |||
AUG | Met | [M] | ACG | Thr | [T] | AAG | Lys | [K] | AGG | Arg | [R] | G | |||
G | GUU | Val | [V] | GCU | Ala | [A] | GAU | Asp | [D] | GGU | Gly | [G] | U | ||
GUC | Val | [V] | GCC | Ala | [A] | GAC | Asp | [D] | GGC | Gly | [G] | C | |||
GUA | Val | [V] | GCA | Ala | [A] | GAA | Glu | [E] | GGA | Gly | [G] | A | |||
GUG | Val | [V] | GCG | Ala | [A] | GAG | Glu | [E] | GGG | Gly | [G] | G | |||
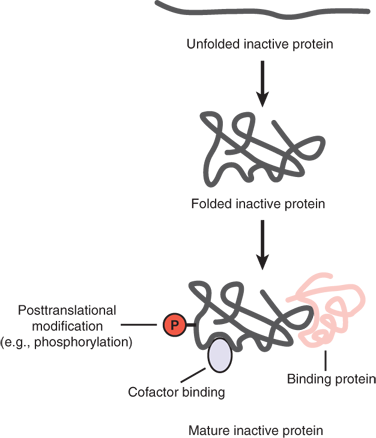
The biologic versatility of proteins is astounding. Among many other functions, proteins serve as enzymes that catalyze critical biochemical reactions, carry signals to and from the extracellular environment, and mediate diverse signaling and regulatory functions in the intracellular environment. They also transport ions and various small molecules across plasma membranes. Proteins make up the key structural components of cells and the extracellular matrix and are responsible for cell motility. The unique functional properties of proteins are largely determined by their structure (Fig. 15-5).
Figure 15-5.
Maturation of a functional protein. Although the linear amino acid sequence of a protein often is shown, the function of a protein also is controlled by its correctly folded three-dimensional structure. In addition, many proteins also have covalent posttranslational modifications such as phosphorylation or noncovalent binding to a small molecule or a protein.
The human organism is made up of a myriad of different cell types that, despite their vastly different characteristics, contain the same genetic material. This cellular diversity is controlled by the genome and accomplished by tight regulation of gene expression. This leads to the synthesis and accumulation of different complements of RNA and, ultimately, to the proteins found in different cell types. For example, muscle and bone express different genes or the same genes at different times. Moreover, the choice of which genes are expressed in a given cell at a given time depends on signals received from its environment. There are multiple levels at which gene expression can be controlled along the pathway from DNA to RNA to protein (see Fig. 15-4). Transcriptional control refers to the mechanism for regulating when and how often a gene is transcribed. Splicing of the primary RNA transcript (RNA processing control) and selection of completed mRNAs for nuclear export (RNA transport control) represent additional potential regulatory steps. The mRNAs in the cytoplasm can be selectively translated by ribosomes (translational control) or selectively stabilized or degraded (mRNA degradation control). Finally, the resulting proteins can undergo selective activation, inactivation, or compartmentalization (protein activity control).
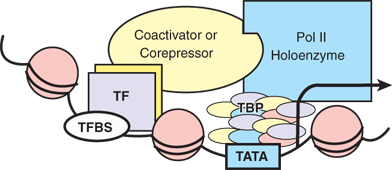
Because a large number of genes are regulated at the transcriptional level, regulation of gene transcripts (i.e., mRNA) often is referred to as gene regulation in a narrow definition. Each of the steps during transcription is properly regulated in eukaryotic cells. Because genes are differentially regulated from one another, one gene can be differentially regulated in different cell types or at different developmental stages. Therefore, gene regulation at the level of transcription is largely context dependent. However, there is a common scheme that applies to transcription at the molecular level (Fig. 15-6). Each gene promoter possesses unique sequences called TATA boxes that can be recognized and bound by a large complex containing RNA polymerase II, forming the basal transcription machinery. Usually located upstream of the TATA box (but sometimes longer distances) are a number of regulatory sequences referred to as enhancers that are recognized by regulatory proteins called transcription factors. These transcription factors specifically bind to the enhancers, often in response to environmental or developmental cues, and cooperate with each other and with basal transcription factors to initiate transcription. Regulatory sequences that negatively regulate the initiation of transcription also are present on the promoter DNA. The transcription factors that bind to these sites are called repressors, in contrast to the activators that activate transcription. The molecular interactions between transcription factors and promoter DNA, as well as between the cooperative transcription factors, are highly regulated and context-dependent. Specifically, the recruitment of transcription factors to the promoter DNA occurs in response to physiologic signals. A number of structural motifs in these DNA-binding transcription factors facilitate this recognition and interaction. These include the helix-turn-helix, the homeodomain motif, the zinc finger, the leucine zipper, and the helix-loop-helix motifs.
Figure 15-6.
Transcriptional control by RNA polymerase. DNA is packaged into a chromatin structure. TATA = the common sequence on the promoter recognized by TBP and polymerase II holoenzyme; TBP = TATA-binding protein and associated factors; TF = hypothetical transcription factor; TFBS = transcription factor binding site; ball-shaped structures = nucleosomes. Coactivator or corepressor is a factor linking the TF with the Pol II complex.
Genome is a collective term for all genes present in one organism. The human genome contains DNA sequences of 3 billion base pairs, carried by 23 pairs of chromosomes. The human genome has an estimated 25,000 to 30,000 genes, and overall, it is 99.9% identical in all people.7,8 Approximately 3 million locations where single-base DNA differences exist have been identified and termed single nucleotide polymorphisms. Single nucleotide polymorphisms may be critical determinants of human variation in disease susceptibility and responses to environmental factors.
The completion of the human genome sequence in 2003 represented another great milestone in modern science. The Human Genome Project created the field of genomics, which is the study of genetic material in detail (see Fig. 15-1). The medical field is building on the knowledge, resources, and technologies emanating from the human genome to further the understanding of the relationship of the genes and their mutations to human health and disease. This expansion of genomics into human health applications resulted in the field of genomic medicine.
The emergence of genomics as a science will transform the practice of medicine and surgery in this century. This breakthrough has allowed scientists the opportunity to gain remarkable insights into the lives of humans. Ultimately, the goal is to use this information to develop new ways to treat, cure, or even prevent the thousands of diseases that afflict humankind. In the twenty-first century, work will begin to incorporate the information embedded in the human genome sequence into surgical practices. By doing so, the genomic information can be used for diagnosing and predicting disease and disease susceptibility. Diagnostic tests can be designed to detect errant genes in patients suspected of having particular diseases or of being at risk for developing them. Furthermore, exploration into the function of each human gene is now possible, which will shed light on how faulty genes play a role in disease causation. This knowledge also makes possible the development of a new generation of therapeutics based on genes. Drug design is being revolutionized as researchers create new classes of medicines based on a reasoned approach to the use of information on gene sequence and protein structure function rather than the traditional trial-and-error method. Drugs targeted to specific sites in the body promise to have fewer side effects than many of today’s medicines. Finally, other applications of genomics will involve the transfer of genes to replace defective versions or the use of gene therapy to enhance normal functions such as immunity.
Proteomics refers to the study of the structure and expression of proteins as well as the interactions among proteins encoded by a human genome (see Fig. 15-1).9 A number of Internet-based repositories for protein sequences exist, including Swiss-Prot (http://www.expasy.ch). These databases allow comparisons of newly identified proteins with previously characterized sequences to allow prediction of similarities, identification of splice variants, and prediction of membrane topology and posttranslational modifications. Tools for proteomic profiling include two-dimensional gel electrophoresis, time-of-flight mass spectrometry, matrix-assisted laser desorption/ionization, and protein microarrays. Structural proteomics aims to describe the three-dimensional structure of proteins that is critical to understanding function. Functional genomics seeks to assign a biochemical, physiologic, cell biologic, and/or developmental function to each predicted gene. An ever-increasing arsenal of approaches, including transgenic animals, RNA interference (RNAi), and various systematic mutational strategies, will allow dissection of functions associated with newly discovered genes. Although the potential of this field of study is vast, it is in its early stages.
It is anticipated that a genomic and proteomic approach to human disease will lead to a new understanding of pathogenesis that will aid in the development of effective strategies for early diagnosis and treatment.10 For example, identification of altered protein expression in organs, cells, subcellular structures, or protein complexes may lead to development of new biomarkers for disease detection. Moreover, improved understanding of how protein structure determines function will allow rational identification of therapeutic targets, and thereby not only accelerate drug development, but also lead to new strategies to evaluate therapeutic efficacy and potential toxicity.9
Every organism is composed of many different cell types at different developmental stages. Some cell types continue to grow, while some cells stop growing after a developmental stage or resume growth after a break. For example, embryonic stem cells grow continuously, while nerve cells and striated muscle cells stop dividing after maturation. Cell cycle is the process for every cell including DNA replication and protein synthesis, DNA segregation in half, and package DNA and protein in two newly formed cells to enable passage of identical genetic information from one parental cell to two daughter cells. Thus, the cell cycle is the fundamental mechanism to maintain tissue homeostasis. A cell cycle comprises four periods: G1 (first gap phase before DNA synthesis), S (synthesis phase when DNA replication occurs), G2 (the gap phase before mitosis), and M (mitosis, the phase when two daughter cells with identical DNA are generated) (Fig. 15-7). After a full cycle, the daughter cells enter G1 again, and when they receive appropriate signals, undergo another cycle, and so on. The machinery that drives cell cycle progression is made up of a group of enzymes called cyclin-dependent kinases (CDKs). Cyclin expression fluctuates during the cell cycle, and cyclins are essential for CDK activities and form complexes with CDK. The cyclin A/CDK1 and cyclin B/CDK1 drive the progression for the M phase, while cyclin A/CDK2 is the primary S phase complex. Early G1 cyclin D/CDK4/6 or late G1 cyclin E/CDK2 controls the G1
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree