Learning Objectives
Introduction
An animal will generally accept an organ transplant from itself, but will reject a transplant from other animals, even if the donor animal is of the same species. Organ rejection is primarily a consequence of the interactions between the immune system of the transplant recipient and the histocompatibility antigens present on the transplanted cells. Clinical laboratories play an important role in the histocompatibility testing for solid organ as well as hematopoietic stem cell and bone marrow transplantation (BMT). This chapter provides a brief background to some of the issues and techniques involved in histocompatibility testing related to transplantation. Other applications of histocompatibility testing such as in the characterization of disease states (eg, HLA-B27 in ankylosing spondylitis) or before initiation of some drug therapy (eg, HLA-B*57:01 is a risk factor for hypersensitivity to abacavir) use similar techniques and are not discussed further.
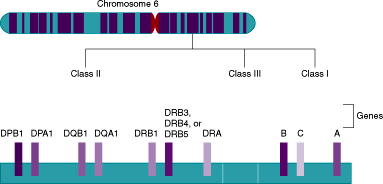
The histocompatibility antigens that are the primary stimulus in graft rejection are encoded by a complex of closely linked genes called the major histocompatibility complex (MHC). In mice, these genes are located on the H2 region of chromosome 17. In humans, the analogous MHC region is located in a 4000-kb region on the short arm of chromosome 6 and encodes for the HLA system (Figure 4–1).
Figure 4–1
Genes of the human MHC system. The human major histocompatibility complex (MHC) is located on the short arm of chromosome 6. It contains over 200 genes that can be divided into different regions (class I-III). Only the major genes encoding the HLA molecules important in transplantation are shown. The class I region contains genes that encode the α chains of the classic transplantation antigens, HLA-A, -B, and -C. The class II region has genes that encode both the α and β chains of HLA-DP, -DQ, and -DR molecules. Genes encoding the α and β chains are designated as “A” or “B,” and are followed by a number if there is more than 1 gene encoding a particular chain or a related pseudogene. For example, DRB1 is 1 of the genes that encodes the β chain of DR molecules. The class III region is located between regions I and II and does not encode for HLA molecules. (Adapted with permission from Clinical Laboratory Reviews [a newsletter publication of the Massachusetts General Hospital] 2000;8:3.)
HLA Genes and Gene Products
The HLA class I region encodes for certain glycoprotein molecules that are present on all nucleated cells. The main function of the HLA class I molecules is to bind to fragments resulting from the breakdown of intracellular pathogens, such as viruses. The HLA molecule and the bound peptide are then presented on the cell surface so that an immune response can be initiated against the pathogen. Class I molecules consist of 2 noncovalently linked chains. A gene in the MHC encodes the heavy chain, and a gene outside the MHC on chromosome 15 encodes the β2-microglobulin light chain. In humans, there are 3 important class I genes known as HLA-A, -B, and -C. These genes are highly polymorphic. Polymorphism refers to multiple variations of a single genetic locus and its gene product. Each variation of the gene is called an allele. The HLA-A, -B, and -C genes encode for over 100 gene products that can be defined by serology—that is, by matching antibodies against the different HLA antigens (Table 4–1). However, the antigens identified by serotyping are far outnumbered by the alleles that are recognized by sequencing the gene. This is because not all gene polymorphisms result in distinct antibody specificities, although they may stimulate a T-cell immune response.
| HLA Gene | Number of Alleles Determined by Gene Sequencinga | Number of Serologic Specificities |
|---|---|---|
| HLA-A | >2200 | 28 |
| HLA-B | >2900 | 70 |
| HLA-C | >1700 | 10 |
| HLA-DRB1 | >1300 | 21 |
| HLA-DQB1 | >320 | 9 |
In humans, there are 3 important class I genes known as HLA-A, -B, and -C. The class II region encodes for the α and β chains that make up the HLA-DR, -DQ, and -DP molecules.
The class II region encodes for the α and β chains that make up the HLA-DR, -DQ, and -DP molecules (Figure 4–1). These HLA class II molecules have a more restricted expression than the class I molecules. They are found mainly on B lymphocytes, dendritic cells, monocytes, activated T cells, and some endothelial cells. Antigen-presenting cells such as macrophages can phagocytose and engulf bacteria and other parasites via endocytosis. Once these pathogens gain entry to the cell, they can be broken down by proteases into peptides that are able to bind to the class II molecules. These peptides are then presented on the cell surface, and an immune response is initiated. The class III region contains approximately 40 genes that do not encode HLA molecules, but encode certain complement components and numerous other proteins not involved in antigen presentation.
The HLA genes are linked together—in what is called a haplotype—on the chromosome and inherited en bloc. Each individual inherits 1 haplotype from each parent, and the 2 haplotypes represent the HLA genotype. The alleles from both of these haplotypes are expressed on an individual’s cells. This is referred to as a codominant expression of the gene. Therefore, even though there are multiple HLA genes, usually only 4 genotypes are possible in the offspring. There is a 25% chance of any 2 siblings being HLA identical and a 50% chance of the siblings sharing a haplotype.
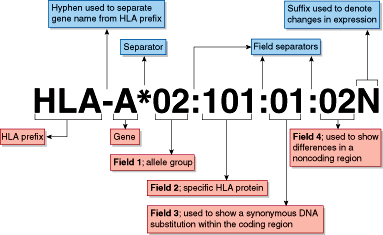
HLA antigens and alleles are named by the World Health Organization Nomenclature Committee for Factors of the HLA System. New alleles are named on an ongoing basis as they are identified, and the number of HLA alleles has grown rapidly, with over 8000 currently listed. The DNA sequences of all recognized HLA alleles are maintained in the IMGT/HLA Database and are available online (http://www.ebi.ac.uk/ipd/imgt/hla/). Each HLA allele name follows a strict format defined by the Nomenclature Committee (Figure 4–2).
Figure 4–2
HLA nomenclature. The letters following the HLA prefix (eg, HLA-A) designate the gene name in the MHC system. An asterisk (*) separates the gene name from the allele name. A low-resolution DNA typing is reported at the level of the allele group with only the first set of digits (eg, HLA-A*02). This is usually the equivalent of a serologic typing result. The specific HLA protein or allele is reported by the second set of digits in a high-resolution typing (eg, HLA-A*02:01). The third set of digits is used only when necessary to indicate synonymous (silent) nucleotide substitutions, and the fourth set of digits is used when necessary to denote substitutions in noncoding regions of the gene. A letter at the end may be used to denote changes in expression (eg, N, null or not expressed). (This figure is kindly provided by Professor Steven G.E. Marsh, Anthony Nolan Research Institute, London, UK, and is from the website hla.alleles.org)
Histocompatibility Testing Assays
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree