severe neutropenia, are specifically at risk of bacteremia caused by these microorganisms (16). Several studies have demonstrated that colonization of the nasopharynx, rectum, or skin by coagulase-negative staphylococci precedes the development of bacteremia, and chemotherapyinduced breaks of the normal mucosa and skin barriers as well as placement of medical devices often represent typical entry sites for the bacteria (17).
a history of a recent surgical procedure. Mortality is high (25%), which is probably also associated with the underlying chronic diseases in these patients (32). In PVE, coagulasenegative staphylococci are isolated as causative pathogens in 15% to 40% of the cases. Infections are usually healthcareassociated and occur within 12 months of valve replacement (33, 34 and 35). Patients present with prolonged symptoms (>1 month) of weakness and low-grade fever. The modified Duke criteria are applied to diagnose infective endocarditis (36), which include, among other factors, positive blood cultures and typical echocardiographic findings.
staphylococcal species is resistant to lysis by lysozyme that normally attacks the β-1,4-glycosidic bonds in the peptidoglycan chains. Lysozyme resistance in staphylococci was found to be based on O-acetylation of the muramic acid of the peptidoglycan, specifically in human pathogens such as S. aureus, S. epidermidis, and Staphylococcus lugdunensis (63,64). Currently, the genus Staphylococcus comprises 47 species and 11 subspecies (Table 30-1). Classification of staphylococci is traditionally based on the production of coagulase, an enzyme that binds fibrinogen and mediates its conversion into fibrin, resulting eventually in blood plasma coagulation. In addition to the major coagulase-positive pathogen S. aureus, six other coagulasepositive species have been described that mainly play a role in veterinary medicine (Table 30-1) (112). Among the 41 coagulase-negative species, S. epidermidis is the most common one with a broad pathogenic potential causing a wide variety of infections. Other coagulase-negative species involved in human disease are Staphylococcus haemolyticus, Staphylococcus saprophyticus, Staphylococcus capitis, S. lugdunensis, Staphylococcus hominis, Staphylococcus warneri, and Staphylococcus schleiferii. The natural reservoir of all staphylococci is the skin and the mucosa of humans and animals where the bacteria mostly reside as harmless and benign commensals. Some species were recovered from processed food or environmental samples and are traditionally used in food industry (e.g., Staphylococcus carnosus, Staphylococcus piscifermentans). In humans, microbial ecology of staphylococcal species varies individually and also depends on the body site. Thus, S. aureus prefers to colonize the anterior nares and the nasopharynx, but only 20% of healthy adults are permanently, and another 60% are transiently colonized by S. aureus (113). In contrast, all human beings are persistently colonized by coagulasenegative staphylococcal species. S. epidermidis is the species that occurs in abundance. It colonizes preferentially the upper part of the body, including the nasopharynx, and constitutes over 50% of the resident staphylococci (114). Other coagulase-negative species have adapted to distinct ecological niches and can be recovered from specific parts of the skin (e.g., S. saprophyticus from the perineum, S. capitis from the scalp, Staphylococcus auricularis from the external ear, S. haemolyticus and S. hominis from the axilla as well as the pubic and the perineal region, respectively).
TABLE 30-1 The Genus Staphylococcus and Its Species | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
initiates with a variety of phenotypic tests, first of all to differentiate between coagulase-negative staphylococci and S. aureus. Slide-agglutination tests are performed to detect S. aureus-specific clumping factor, and other cell wall-associated proteins of S. aureus. As slide agglutination occasionally provides ambiguous results, the classical coagulase test mentioned above may be performed as well. Putative coagulase-negative staphylococcal species are then further examined for their colony morphology, growth requirements, oxidative and fermentative utilization of carbohydrates, novobiocin susceptibility, and various enzymatic activities (e.g., nitrate reductase, alkaline phosphatase, urease, ornithine decarboxylase, arginine dehydrogenase, etc.) (61). A variety of commercial test kits such as the API 20 Staph and API ID32 Staph systems (bioMerieux), the Staph-Zym (Rosco) or the Vitek system (bioMerieux) are available that combine detection of most of these phenotypic properties and allow for a rapid and convenient identification in the routine diagnostic laboratory. However, phenotypic tests have an inherent weakness as they rely on the expression of the phenotypic characteristic in question, and in coagulase-negative staphylococci these properties may vary within isolates belonging to the same species (115,116,117). Thus, DNA-based genotyping methods become increasingly important for an expression-independent species identification of coagulase-negative staphylococci (118). These methods target highly conserved species-specific DNA loci and genes that are PCR-amplified and subjected to either DNA-fragment pattern comparison or DNA sequencing. PCR-based approaches include ribotyping, amplified-fragment-lengthpolymorphism (AFLP), and tRNA-locus-interspacer-lengthpolymorphism (tDNA-ILP) analyses (Table 30-2). DNA sequencing of amplified 16S rRNA DNA loci is the most common method for species identification across many bacterial genera (122). Due to the close relatedness of some coagulase-negative staphylococcal species, the method may not have, in all cases, sufficient discriminatory power (126). Therefore, DNA sequencing of a range of housekeeping genes has been implemented to complement 16S rRNA locus analysis (see Table 30-2 for details and references). In general, genotyping methods are considered to be superior to mere phenotyping for coagulase-negative Staphylococcus species definition. With the increasing availability and cost effectiveness of molecular techniques in diagnostic laboratories, accurate species identification of coagulase-negative staphylococci can be anticipated to become a routine procedure. This most welcome development in microbiological diagnostics will surely shed more light on the association of specific coagulase-negative staphylococcal species with distinct infection processes.
TABLE 30-2 Genotypic Methods for Species Identification of Coagulase-Negative Staphylococcal Species | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
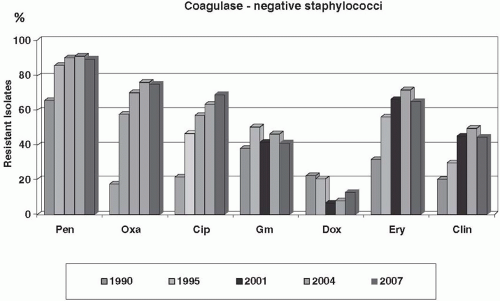
resistance emerged very soon, and resistance rates are nowadays as high as 70% to 90% among coagulase-negative staphylococci. Methicillin resistance is mediated by the mecA gene complex that is located on a unique molecular vector called the staphylococcal chromosome cassette (SCCmec) (133). mecA encodes the additional low-affinity penicillin-binding protein PBP2a that enables cell wall synthesis in the presence of β-lactam antibiotics. SCCmec cassettes represent large chromosomal DNA fragments that may harbor, in addition to mecA, a great variety of accessory genes, for example restriction modification systems, metabolic genes, integrated plasmids, transposons, insertion sequence (IS) elements, and many more. A characteristic feature of SCCmec cassettes is the presence of recombinase genes that confer mobility and mediate the site-specific integration of the elements into a highly conserved locus of the Staphylococcus chromosome (i.e., orfX). SCCmecs have an independent evolutionary history and eight major SCCmec types have been described to date (133,134). They are considered to be transferred into S. aureus from a coagulase-negative species (135,136). However, the evolutionary origin of SSCmecs, the mechanism of SCCmec acquisition, and the factors that favor or limit their spread are still poorly understood. Interestingly, SCC cassettes cannot only carry mecA and mediate methicillin resistance. A number of SCCs has been identified that are devoid of mecA, but carry other genes instead (135,137). Therefore, SCCs are regarded as effective vectors to spread useful genes among staphylococci (138).
phenotype depends on the expression status of the respective erm genes. In staphylococci, ermA and ermC can be expressed either constitutively or inducibly (144,145). Constitutive expression results in cross-resistance against all three antibiotic classes. In contrast, strains with inducible erm expression display in vitro resistance to 14- and 15-membered ring macrolides, which also represent inducer molecules, while retaining susceptibility toward clindamycin and group B streptogramins. Although clindamycin is not able to induce ermA or ermC expression directly, exposure of erm-inducible staphylococcal isolates to clindamycin may result in complete MLSB cross resistance both in vitro and in vivo. This phenomenon is attributed to the selection of preexisting constitutive erm mutants (144). As clindamycin is an alternative drug for the treatment of some staphylococcal infections, detection of the inducible MLSB resistance phenotype is of clinical relevance and should be performed when required by the D-test (Fig. 30-2) (146).
|
has been reported for a number of other coagulase-negative staphylococcal species as well (e.g., S. epidermidis, S. warneri), and it is therefore necessary to perform careful antibiotic resistance testing to avoid treatment failure (153,155,159,160).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree