Key Points
Summary:
Pregnancies found to be at greater than population (high) risk for a genetic disorder are offered invasive diagnostic testing by chorionic villus sampling (CVS) or amniocentesis. These diagnostic tests have a small risk of inducing a pregnancy loss but will diagnose a specific genetic disorder with greater than 99% accuracy.
Karyotype, FISH, or microarray are testing options for diagnosis of fetal genetic disorders on chorionic villus or amniotic fluid specimens.
Ancestry and family history are tools used to determine which pregnancies are at risk for specific genetic disorders and should help guide carrier screening.
The carrier state of some Mendelian disorders is frequent enough in the general population that offering screening to all pregnant couples and those presenting preconception is recommended.
Screening tests can be used during pregnancy, at as early as 10 weeks, to identify pregnancies at increased risk for common fetal trisomies such as those causing Down syndrome and Edwards syndrome (trisomy 21 and 18).
Uses:
Invasive testing—CVS and amniocentesis
Provide a definitive diagnosis for pregnancies identified as having an increased risk of a genetic disorder.
Invasive testing is most commonly performed on women who are at an increased risk for a chromosome abnormality based on age-related risk, abnormal noninvasive screening results, and/or abnormal ultrasound findings.
Invasive testing is offered for disease-specific testing when the parent(s) are confirmed carrier(s), affected with a dominant genetic disorder, or known to carry a balanced translocation.
American College of Obstetrics and Gynecology, Guidelines, 2007
All women, regardless of age, should have the option of invasive testing.
Maternal age of 35 years alone should no longer be used as a cutoff to determine who is offered screening versus who is offered invasive testing.
General Descriptions
Purpose: to identify carrier couples of Mendelian disorders
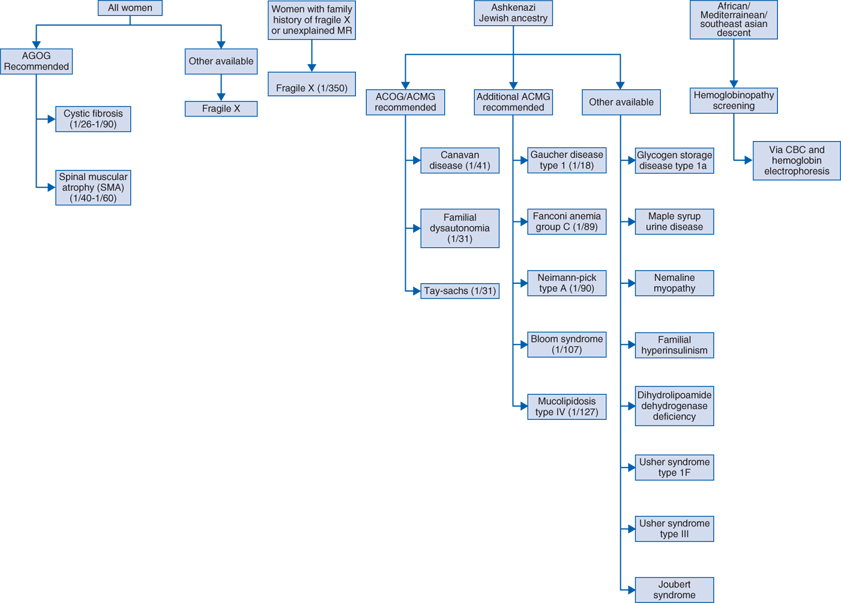
Population screening for carriers of common Mendelian disorders
Some disorders occur frequently enough throughout the general population that carrier testing is offered to all pregnant couples, for example, cystic fibrosis (Table 98-1).
Population screening based on ethnicity and race
The carrier state of certain genetic disorders occurs at an increased rate in specific ethnic or racial populations (Table 98-1). Since many of these disorders are inherited in an autosomal recessive fashion, identification of couples in which both parents are phenotypically normal but carry a mutated gene for the same disorder allows early decisions about future reproduction.
Preferably, carrier screening should be performed preconception or as early in pregnancy as possible. Turnaround time averages are 1 to 2 weeks.
Carrier screening is usually performed in a sequential manor—that is, if the mother tests positive, then the father of the pregnancy is tested. The exception to this is when time constraints, require a rapid analysis of both partners. If the parents are of different ethnicities, the father can be offered screening for those disorders not recommended in the mother based on her ethnicity.
Screening couples based on family history
Specific testing for disorder found in family history (ie, cystic fibrosis, Duchenne muscular dystrophy).
Testing can be targeted to look for specific disease mutation found in family member if known.
If it is determined that a fetus is at risk based on carrier screening results, the patient should be offered CVS or amniocentesis to test for that specific disorder.
Purpose: 1. To identify pregnancies at increased risk for common aneuploidies or genetic disorders
Prenatal maternal serum screening tests and ultrasound are used to target women at high risk (> general population) for fetal aneuploidy (especially Down syndrome and trisomies 13 and 18) and birth defects. Women found to be at high risk can then be offered invasive diagnostic testing.
Abnormal maternal serum screening results may also indicate an increased risk for an adverse pregnancy (eg, spontaneous abortion, preterm delivery, low birth weight), or other genetic syndromes.
Most women who have an abnormal first- or second-trimester maternal serum screening will have normal pregnancies.
Certain ultrasound anomalies are associated with specific genetic disorders and if found, patients should be offered testing for that disorder (ie, testing for DiGeorge syndrome in a patient with a heart defect).
Purpose: 2. To avoid the conception of a pregnancy with a genetic disorder
Preimplantation genetic diagnosis (PGD)—for some couples at high risk to have a pregnancy affected with a genetic disorder, PGD may be performed so that only genetically unaffected embryos are implanted. In this approach, in vitro fertilization (IVF) is required and one or two cells (blastomeres) are removed at either day 3 or day 5 postfertilization. Single cell analysis is then performed and results may be available with 24 to 48 hours. Because PGD is not perfect, invasive testing should be offered as confirmation.
Chorionic villus sampling (CVS)
Performed between 10 and 12 to 14 weeks’ gestation (depending on comfort level or experience of the technician)
CVS can be performed abdominally or cervically, depending on the location of the placenta.
During the procedure, a needle (abdominally) or catheter (cervically) is inserted into the placenta and some of the cells that make up the placenta, called the chorionic villi, are removed.
CVS cannot detect open neural tube defects (ONTDs). Therefore, in the event a CVS is performed, a second-trimester maternal alpha-fetoprotein (AFP) blood test should be performed and along with an anatomy scan, as together they can detect 90% of ONTDs.
Amniocentesis
Is performed beginning at 15 to 16 weeks’ gestation and can be performed until delivery.
Amniocentesis is performed abdominally.
During the procedure, a needle is inserted into the abdomen and some of the amniotic fluid is removed.
Amniocentesis can detect ONTDs with a 95% detection rate.
Laboratory testing of CVS or amniocentesis samples
Karyotype
Looks at the chromosomes under a light microscope and uses the banding pattern of the chromosomes to determine if there are aberrations. It is able to detect deletions or duplications greater than 5 to 10 MB in size.
Is the gold standard testing performed on all CVS and amniocentesis samples.
Turnaround time averages 7 to 10 days.
Fluorescence in situ hybridization testing (FISH)
Uses fluorescent probes that attach to specific regions of the genome (ie, chromosome 21 or region 22q11.2) to determine copy number of that particular chromosome or region.
Is able to detect targeted smaller deletions or duplications than karyotype can, but you have to know exactly what you are looking for (which region to test).
Commonly used to provide quick results for the common aneuploidies (13, 18, 21, X, and Y) as turnaround time is 1 to 3 days.
Also used when testing for a specific microdeletion or duplication syndrome is warranted, such as testing for 22q11.2 deletion in the presence of a heart defect.

Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree