3.1 INTRODUCTION TO VIRUS STRUCTURE
Outside their host cells, viruses survive as virus particles, also known as virions. The virion is a gene delivery system; it contains the virus genome, and its functions are to protect the genome and to aid its entry into a host cell, where it can be replicated and packaged into new virions. The genome is packaged in a protein structure known as a capsid.
Many viruses also have a lipid component, generally present at the surface of the virion, forming an envelope which also contains proteins that play a role in aiding entry into host cells. A few viruses form protective protein occlusion bodies around their virions. Before looking at these virus structures we shall consider characteristics of the nucleic acid and protein molecules that are the main components of virions.
3.2 VIRUS GENOMES
A virion contains the genome of a virus in the form of one or more molecules of nucleic acid. For any one virus the genome is composed of either RNA or DNA. If a new virus is isolated, one way to determine whether it is an RNA virus or a DNA virus is to test its susceptibility to a ribonuclease and a deoxyribonuclease. The virus nucleic acid will be susceptible to degradation by only one of these enzymes.
Each nucleic acid molecule is either single-stranded (ss) or double-stranded (ds), giving four categories of virus genome: dsDNA, ssDNA, dsRNA, and ssRNA. The dsDNA viruses encode their genes in the same kind of molecule as animals, plants, bacteria, and other cellular organisms, while the other three types of genome are unique to viruses. It interesting to note that most fungal viruses have dsRNA genomes, most plant viruses have ssRNA genomes, and most prokaryotic viruses have dsDNA genomes. The reasons for these distributions presumably concern diverse origins of the viruses in these very different host types.
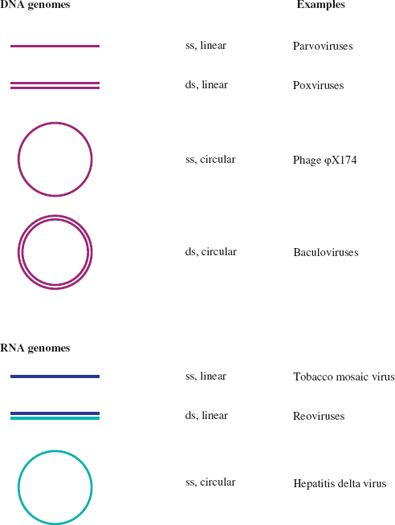
A further categorization of a virus nucleic acid can be made on the basis of whether the molecule is linear, with free 5′ and 3′ ends, or circular, as a result of the strand(s) being covalently closed. Examples of each category are given in Figure 3.1. In this figure, and indeed throughout the book, molecules of DNA and RNA are color coded.
Figure 3.1 Linear and circular viral genomes.
ss: single-stranded
ds: double-stranded
Dark blue and light blue depict (+) RNA and (−) RNA respectively; these terms are explained in Section 6.2.
There are no viruses known with circular dsRNA genomes.
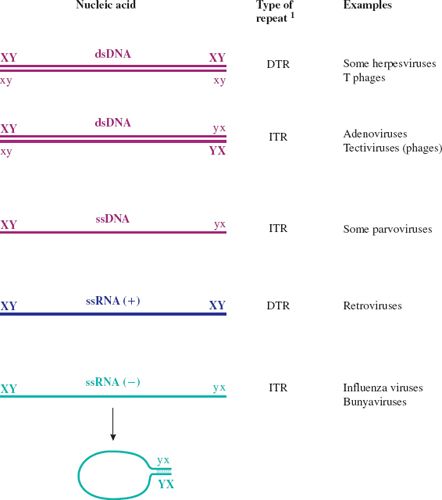
It should be noted that some linear molecules may be in a circular conformation as a result of base pairing between complementary sequences at their ends (see Figure 3.7). This applies, for example, to the DNA in hepadnavirus virions and to the RNA in influenza virions.
Figure 3.7 Terminal repeats in virus genomes.
1 DTR: direct terminal repeat.
ITR: inverted terminal repeat.
X and x represent complementary sequences.
Y and y represent complementary sequences.
ssRNA (+) has the same sequence as the virus mRNA.
ssRNA (–) has the sequence complementary to the virus mRNA.
The RNAs of ssRNA viruses with ITRs can circularize; a “panhandle” is formed by base pairing between the complementary sequences at the termini.
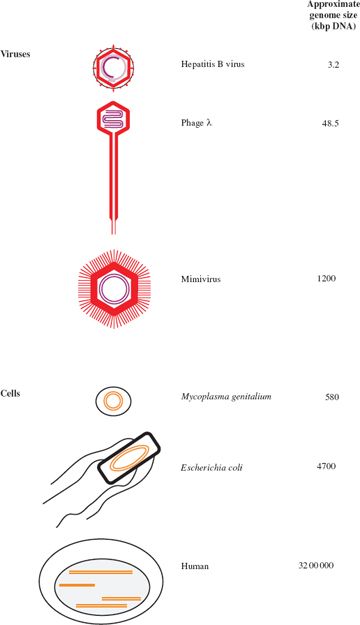
3.2.1 Genome size
Virus genomes span a large range of sizes. The smallest virus known is tobacco necrosis satellite virus, with a ssRNA genome of 1239 bases, while at the other end of the scale there are viruses with dsDNA genomes of over 1000 kilobase pairs (kbp). The largest virus genomes, such as that of the mimivirus, are larger than the smallest genomes of cellular organisms, such as some mycoplasmas (Figure 3.2).
The maximum size of a virus genome is subject to constraints, which vary with the genome category. As the constraints are less severe for dsDNA all of the large virus genomes are composed of dsDNA. The genomes of some dsDNA viruses have been found to encode small RNAs (transfer RNAs and micro-RNAs), as well as proteins. The largest RNA genomes known are those of some coronaviruses, which are 33 kb of ssRNA.
Viruses with small genomes have evolved economical ways of using their limited coding capacities. Some viruses, such as hepatitis B virus (Section 19.6), use every nucleotide for protein coding and encode proteins in overlapping reading frames. Another way that small viruses have circumvented the limitation of a small genome is by evolving proteins that can perform two or more functions (Section 3.3).
3.2.2 Secondary and tertiary structure
As well as encoding the virus proteins, and in some cases RNAs, to be synthesized in the infected cell, the virus genome carries additional information, such as signals for the control of gene expression. Some of this information is contained within the nucleotide sequences, while for the single-stranded genomes some of it is contained within structures formed by intramolecular base pairing.
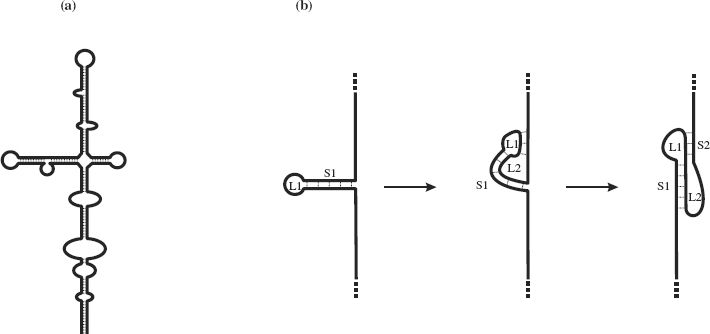
In ssDNA, complementary sequences may base pair through G—C and A—T hydrogen bonding; in ssRNA weaker G—U bonds may form in addition to G—C and A—U base pairing. Intramolecular base pairing results in regions of secondary structure with stem-loops and bulges (Figure 3.3(a)). In some ss-RNAs, intramolecular base pairing results in structures known as pseudoknots, the simplest form of which is depicted in Figure 3.3(b). Some pseudoknots have enzyme activity, while others play a role in ribosomal frameshifting (Section 6.4.2).
Figure 3.3 Secondary structures resulting from intramolecular base-pairing in single-stranded nucleic acids. (a) Stem-loops and bulges in ssRNA and ssDNA. (b) Formation of a pseudoknot in ssRNA. A pseudoknot is formed when a sequence in a loop (L1) at the end of a stem (S1) base-pairs with a complementary sequence outside the loop. This forms a second loop (L2) and a second stem (S2).
Regions of secondary structure in single-stranded nucleic acids are folded into tertiary structures with specific shapes, many of which are important in molecular interactions during virus replication. For an example see Figure 14.5, which depicts the 5′ end of poliovirus RNA, where there is an internal ribosome entry site to which cell proteins bind to initiate translation. Another example can be seen in Figure 18.3, which depicts the 5′ end of HIV-1 RNA, where there are a number of regions with specific functions.
3.2.3 Modifications at the ends of virus genomes
It is interesting to note that the genomes of some DNA viruses and many RNA viruses are modified at one or both ends (Figures 3.4 and 3.5). Some genomes have a covalently linked protein at the 5′ end. In at least some viruses this is a vestige of a primer that was used for initiation of genome synthesis (Section 7.3.1).
Figure 3.4 DNA virus genomes with one or both ends modified. The 5′ end of some DNAs is covalently linked to a protein. One of the hepatitis B virus DNA strands (the (+) strand) is linked to a short sequence of RNA with a methylated nucleotide cap.
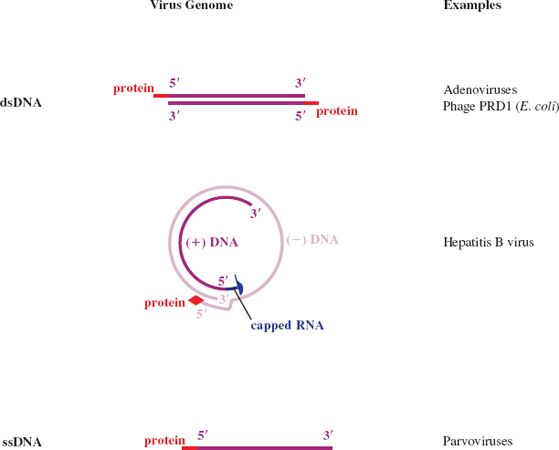
Figure 3.5 RNA virus genomes with one or both ends modified. The 5′ end may be linked to a protein or a methylated nucleotide cap. The 3′ end may be polyadenylated or it may be folded like a transfer RNA.
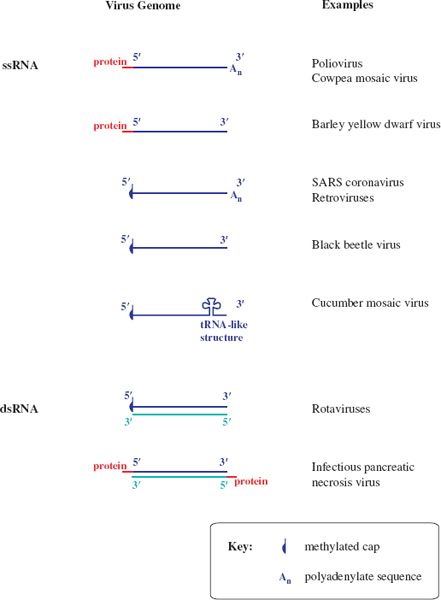
Some genome RNAs have one or both of the modifications that occur in eukaryotic messenger RNAs (mRNAs): a methylated nucleotide cap at the 5′ end (Section 6.3.4) and a sequence of adenosine residues (a polyadenylate tail; poly(A) tail) at the 3′ end (Section 6.3.5).
The genomes of many ssRNA viruses, including most of those in Figure 3.5, function as mRNAs after they have infected host cells. It is worth noting that very few of these RNAs have both a cap and a poly(A) tail, and some have neither.
The genomes of some ssRNA plant viruses are base paired and folded near their 3′ ends to form structures similar to transfer RNA. These structures contain sequences that promote the initiation of RNA synthesis.
3.2.4 Proteins non-covalently associated with virus genomes
Many nucleic acids packaged in virions have proteins bound to them non-covalently. These proteins have regions that are rich in the basic amino acids lysine, arginine, and histidine, which are negatively charged and able to bind strongly to the positively charged nucleic acids.
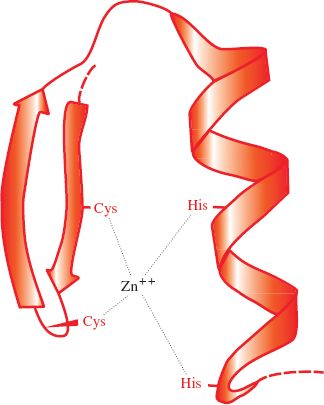
Papillomaviruses and polyomaviruses, which are DNA viruses, have cell histones bound to the virus genome. Most proteins associated with virus genomes, however, are virus coded, such as the HIV-1 nucleocapsid protein that coats the virus RNA; 29% of its amino acid residues are basic. As well as their basic nature, nucleic acid-binding proteins may have other characteristics, such as zinc fingers (Figure 3.6); the HIV-1 nucleocapsid protein has two zinc fingers.
Figure 3.6 A zinc finger in a protein molecule. A zinc finger has recurring cysteine and/or histidine residues at regular intervals. In this example there are two cysteines and two histidines.
In some viruses, such as tobacco mosaic virus (Section 3.4.1), the protein coating the genome constitutes the capsid of the virion.
3.2.5 Segmented genomes
Most virus genomes consist of a single molecule of nucleic acid, but the genes of some viruses are encoded in two or more nucleic acid molecules. These segmented genomes are much more common amongst RNA viruses than DNA viruses. Examples of ssRNA viruses with segmented genomes are the influenza viruses (Chapter 16), which package the segments in one virion, and brome mosaic virus, which packages the segments in separate virions. Most dsRNA viruses, such as members of the family Reoviridae (Chapter 13), have segmented genomes.
The possession of a segmented genome provides a virus with the possibility of new gene combinations, and hence a potential for more rapid evolution (Section 20.3.3.c). There are, however, prices to be paid for this advantage. Viruses with the genome segments packaged in one virion need an efficient mechanism to ensure that each virion packages a full set of segments. For viruses with the segments packaged in separate virions at least one of each virion category must enter a cell in order to initiate an infection.
3.2.6 Repeat sequences
The genomes of many viruses contain sequences that are repeated. These sequences include promoters, enhancers, origins of replication, and other elements that are involved in the control of events in virus replication. Many linear virus genomes have repeat sequences at the ends (termini), in which case the sequences are known as terminal repeats (Figure 3.7). If the repeats are in the same orientation they are known as direct terminal repeats (DTRs), whereas if they are in the opposite orientation they are known as inverted terminal repeats (ITRs). Strictly speaking, the sequences referred to as “ITRs” in single-stranded nucleic acids are not repeats unless the molecule becomes double stranded during replication. These “ITRs” are, in fact, repeats of the complementary sequences (see ssDNA and ssRNA (–) in Figure 3.7).
3.3 VIRUS PROTEINS
The virion of tobacco mosaic virus contains only one protein species and the virions of parvoviruses contain two to four protein species. These are viruses with small genomes. As the size of the genome increases, so the number of protein species tends to increase; 50 protein species have been reported in the virion of herpes simplex virus 1, and over 140 in the virion of the algal virus Paramecium bursaria Chlorella virus 1.
Proteins that are components of virions are known as structural proteins. They have to carry out a wide range of functions, including:
- protection of the virus genome;
- attachment of the virion to a host cell (for many viruses);
- fusion of the virion envelope to a cell membrane (for enveloped viruses).
Virus proteins may have additional roles, some of which may be carried out by structural proteins, and some by non-structural proteins (proteins which are synthesized by the virus in an infected cell but which are not virion components). These additional roles include:
- enzymes, e.g. protease, reverse transcriptase;
- transcription factors;
- primers for nucleic acid replication;
- formation of ion channels in viral and cell membranes;
- interference with the immune response of the host.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree